Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista Pan-Amazônica de Saúde
versão impressa ISSN 2176-6215versão On-line ISSN 2176-6223
Rev Pan-Amaz Saude v.1 n.2 Ananindeua jun. 2010
http://dx.doi.org/10.5123/S2176-62232010000200005
ARTIGO ORIGINAL | ORIGINAL ARTICLE | ARTÍCULO ORIGINAL
Comparison of two DNA obtainment methods as alternative protocols for the detection of human malaria parasites by nested PCR
Comparação entre dois métodos de obtenção de DNA a serem usados como protocolos alternativos para a detecção de parasitas humanos causadores de malária por nested PCR
Comparación entre dos métodos de obtención de DNA a ser usados como protocolos alternativos para la detección de parásitos humanos causadores de malaria por nested PCR
Giselle Maria Rachid Viana; Danielle Regina Lima Barbosa; Ediclei Lima do Carmo; José Mário Veloso Peres; José Maria Souza Nascimento; Marinete Marins Póvoa
Laboratório de Pesquisas Básicas em Malária, Seção de Parasitologia, Instituto Evandro Chagas/SVS/MS, Ananindeua, Pará, Brasil
Endereço para correspondência
Correspondence
Dirección para correspondencia
ABSTRACT
The correct and precise laboratory diagnosis of human malaria is still a challenge because the reference method, the Giemsa-stained thick blood smear (TS), has limitations that present problems for malaria control. Because of these problems, several studies have attempted to develop alternative methods for malaria diagnosis. Many of these studies focus on molecular diagnosis methods and have led to the development of some alternatives to TS. However, their limitations include high cost, protocol complexity and variable quality of DNA sources and reagents. Nested PCR has been shown to be a good method in this respect and it can be improved by using a high-quality source of DNA. In this study we evaluated two methods for the obtainment of DNA from dried blood samples on filter paper: 1) washing and 2) saponin/chelex-100. The second method showed higher sensitivity and specificity compared to the first, as it detected more infections, whether single or mixed, as well as Plasmodium malariae infections. Based on these results, we present this method as the protocol of choice for DNA obtainment. Nested PCR using saponin/chelex-100 for DNA extraction could be an alternative or complementary diagnosis method for human malaria parasites, but it is not appropriate for routine use.
Keywords: Malaria; DNA; Polymerase Chain Reaction.
RESUMO
Um diagnóstico laboratorial correto e preciso de malária humana ainda é considerado um desafio, pois o método de referência, o da gota espessa com colocação pelo Giemsa, apresenta limitações que dificultam o controle da malária. Devido a esses problemas, várias pesquisas têm objetivado desenvolver métodos alternativos para o diagnóstico da malária. Grande parte desses estudos aborda métodos de diagnóstico molecular, que têm acarretado o desenvolvimento de algumas alternativas ao método de coloração pelo Giemsa. No entanto, esses métodos, por sua vez, apresentam suas limitações, que incluem seu alto custo, a complexidade do protocolo e a variação da qualidade das fontes de DNA e de reagentes. Neste aspecto, a técnica de nested PCR tem demonstrado ser um bom método e pode ser melhorado usando uma fonte de DNA de alta qualidade. Neste estudo foram avaliados dois métodos para a obtenção de DNA de amostras de sangue seco colhidas em papel de filtro: 1) lavagem e 2) saponina/Chelex-100. O segundo método apresentou sensibilidade e especificidade mais altas em relação ao primeiro, pois detectou mais infecções, tanto simples como mistas, bem como infecções por Plasmodium malariae. Com base nesses resultados, apresentamos o segundo como o protocolo de escolha para a obtenção de DNA. A técnica de nested PCR usando saponina/Chelex-100 para extração de DNA pode ser um método alternativo ou complementar de diagnóstico de parasitas da malária humana, mas não é considerado adequado para o uso de rotina.
Palavras-chave: Malária; DNA; Reação em Cadeia da Polimerase.
RESUMEN
Un diagnóstico de laboratorio correcto y preciso de malaria humana todavia se considera un desafio, pues el método de referencia, el de la gota espesa con colocación en Giemsa, presenta limitaciones que dificultan el control de la malaria. Debido a esos problemas, varias investigaciones han tenido como objetivo desarrollar métodos alternativos para el diagnóstico de la malaria. Gran parte de esos estudios aborda métodos de diagnóstico molecular, que han resultado en el desarrollo de algunas alternativas al método de tenido por Giemsa. Sin embargo, esos métodos, por su vez, presentan limitaciones, que incluyen su alto costo, la complejidad del protocolo y la variación de la calidad de las fuentes de DNA y de reactivos. En este aspecto, la técnica de nested PCR ha demostrado ser un buen método y puede ser mejorado usando una fuente de DNA de alta calidad. En este estudio se evaluaron dos métodos para la obtención de DNA de muestras de sangre seca recogidas en papel de filtro: 1) lavado y 2) saponina/Chelex-100. El segundo método presentó sensibilidad y especificidad más altas en relación al primero, pues detectó más infecciones, tanto simples como mixtas, bien como infecciones por Plasmodium malariae. Con base en estos resultados, presentamos el segundo método como el protocolo de elección para la obtención de DNA. La técnica de nested PCR usando saponina/Chelex-100 para extracción de DNA puede ser un método alternativo o complementario de diagnóstico de parásitos de la malaria humana, pero no se considera adecuado para uso de rutina.
Palabras clave: Malaria; ADN; Reacción en Cadena de la Polimerasa.
INTRODUCTION
Malaria is one of the main public health problems in Brazil, affecting mainly poor populations in the Amazon region16,18. The precise identification of the human-infecting Plasmodium species is very important to ensure adequate and early treatment. The reference method for malaria diagnosis is the Giemsa-stained thick blood smear (TS); it is inexpensive, has good sensitivity and specificity and allows for the identification of all developing forms of the Plasmodium species as well as the quantification of parasite stages8,21. Generally, the sensitivity range for detection by microscopists is 10-30 parasites/µL of blood; however, sensitivity can be limited when there is a high volume of samples to be examined in a very short time. It is possible to obtain false-negative results due to low levels of parasitemia, mixed infections or inexperience on the part of the microscopist. This fact can compromise one of the main strategies for malaria control: early and precise diagnosis for the implementation of the correct treatment2,3,7,20.
In order to overcome some of the limitations of TS, methods based on the Polymerase Chain Reaction (PCR), such as the nested PCR protocol developed by Kimura et al11, have been used to detect and identify malaria parasites. These methods are more sensitive and specific than TS and can detect even a single parasite per microliter of blood5,10,21,23. However, the success of a PCR technique depends on various factors, such as the quality of the DNA obtained from different sources and of the reagents. In addition, it has been demonstrated that DNA amplification from dried blood samples on filter paper is a reliable source of DNA when the transportation conditions, manipulation and packing are adequate to prevent DNA contamination or degradation1,4,6,12. For this reason, it is important to test different DNA obtainment methods as alternative protocols for the detection of human malaria parasites by nested PCR. This study aimed to compare the two following techniques: 1) DNA extraction by washing26 and 2) DNA extraction by saponin/chelex-100 treatment of dried blood samples on filter paper27.
MATERIALS AND METHODS
In this study we used 75 positive (range of parasitemia – 0.001% to 2%) and 78 negative dried blood samples on 2.5-cm-diameter Whatman® filter paper discs (Titertek, ICN Biomedicals – England). Blood samples were collected from people living in the municipalities of Novo Repartimento (04o14' 55'' N; 50o 07' 25'' W), Parauapebas (06o 04' 03" N; 49o 54' 08" W), Tucuruí (03o 45' 03" N; 49o 40' 03" W) and Belém (01o 27' 20" N; 48o 30' 15" W), in the State of Pará. Controls for each human malaria parasite included well characterized positive samples and ultrapure water and human DNA for the negative controls. All samples were examined by Giemsa-stained thick blood smear (TS) for diagnosis. Quantification of parasites was carried out for the positive slides14.
DNA OBTAINED BY THE WASHING METHOD
The filter paper samples were prepared with five drops of 20 µL of each sample on Whatman® filter paper discs (2.5-cm diameter) (Titertek, ICN Biomedicals – England) and dried at room temperature (RT, 25-27o C). The samples were then packed into pre-labeled plastic bags (BHL Limited, Poole – England) that were kept at -20o C until needed. They were washed with sterile distilled water and 0.9% saline solution using a Millipore® filter system to remove all PCR inhibitors26.
DNA EXTRACTION BY THE SAPONIN/CHELEX-100 METHOD
One-quarter of each dried blood spot on filter paper was cut into small pieces and incubated with 0.5% saponin solution in PBS in an ice bath for one hour. Afterwards, the solution was mixed using a vortex and the supernatant was discarded. This was followed by a wash with PBS, the addition of 20% chelex-100 solution in water and incubation in a dry bath for one hour. After the incubation, the supernatant was transferred to sterile Eppendorf® tubes and kept at -20o C27.
NESTED PCR REACTION
The reaction for the amplification of the 18S gene subunit of the Plasmodium rRNA (ssu-rRNA) was conducted according to the protocol of Kimura et al11. Primers P1 and P2 were used as universal primers in the first round and primers F2, M1 and V1 were used as specific primers for P. falciparum, P. malariae and P. vivax, respectively, in the second round. The nested PCR products obtained were fractionated electrophoretically in 2% agarose gel (Ultra pure agarose, BRL 155517-014) at 100 V for one hour and stained with ethidium bromide (5 µL/mL). The expected PCR product sizes were 130 bp for the first step and 100-110 bp for the second step. Technicians conducting the nested PCR procedure were blinded to any microscopy results.
ETHICS
The protocol of this study was approved by the Research Ethics Committee of the Instituto Evandro Chagas (004/2003-CEP/IEC/SVS/MS). The purpose and procedures of the study were explained to all participants, signature on a written informed consent form was obtained from all patients and socio-demographic and clinical data were collected.
RESULTS
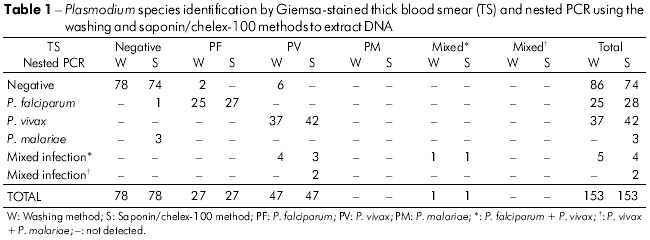
Out of 153 samples, 49.01% (75/153) were positive by TS (parasitemia range 0.001% to 2.0%) and 50.98% (78/153) were negative. With nested PCR, 43.79% (67/153) of samples were positive and 56.20% (86/153) were found to be negative with the washing method for DNA extraction, compared to 51.63% (79/153) positive and 48.36% (74/153) negative with the saponin-chelex-100 method (Table 1).
The performance of nested PCR compared to TS was as follows: 1) washing method - sensitivity = 89.33%, specificity = 100%, accuracy = 94.77%; and 2) saponin/chelex-100 method - sensitivity = 100%, specificity = 94.87%, accuracy = 97.39%. The agreement between these methods and microscopic examination was almost perfect [Kappa = 0.8952 and 0.9477, respectively; CI range: 0.80 to 1.00]. The saponin/chelex-100 method provides a higher quality DNA sample than is obtained by the washing method.
The frequency of Plasmodium species and mixed infections detected by nested PCR using DNA extracted by the washing and saponin/chelex-100 methods is shown in table 2. The saponin/chelex-100 method provided better DNA samples, as demonstrated by the fact that, with this method, nested PCR could detect P. malariae infections either alone or in mixed infection and detected a greater number of other infections compared to the washing method.

Figure 1 shows the results of a nested PCR analysis using DNA obtained by the saponin-chelex-100 method. Lanes 2, 3 and 4 contain samples from a single patient; each well corresponds to reactions for P. falciparum, P. malariae and P. vivax respectively, showing that the patient has a mixed infection with P. malariae and P. vivax.

DISCUSSION
Although the Giemsa-stained thick blood smear (TS) remains the reference method for malaria diagnosis, molecular techniques are more sensitive and specific in detecting parasites and they can be used to evaluate the performance of microscopy20,28.
Dried blood samples on filter paper provide a simple and feasible method for collection and storage and they have be widely adopted in diagnostic screening, genetic analysis and molecular epidemiologic studies in remote areas with tropical climates where transport and storage conditions are often not optimal. Loss of sensitivity of PCR-based methods due to field conditions has already been reported and it is probably a consequence of lower purity, stability and integrity of the DNA obtained from blood samples on filter paper. A problem with sample collection on filter paper is the limitation in the sample volume that can be used for DNA extraction. Therefore, an optimal DNA isolation process is a prerequisite to ensuring the quality and efficiency of molecular methods7,20.
Several studies have been aimed at decreasing the costs of PCR-based protocols and making them rapid and accurate. One part of the protocol that is crucial for getting good results is the obtainment of a high-quality DNA sample1,5,6,7.
In this study, we carried out a comparative analysis of two protocols for DNA obtainment, taking into account the time requirement, the cost and the quality of the extracted DNA7. The results showed that the nested PCR/washing method failed to detect eight positive samples (six P. vivax and two P. falciparum) identified by TS whereas the nested PCR/saponin/chelex-100 method detected all positive samples detected by TS, as well as an additional five samples (three P. malariae single infections and two mixed infections with P. vivax and P. malariae). One possible reason for the failure to detect some positive samples is that the washing method did not properly eliminate nested PCR inhibitors such as hemoglobin. Results showed that the nested PCR/saponin/chelex-100 method was more sensitive and specific than the washing method, especially for the detection of P. malariae.
Analyses of DNA extraction by washing as described by Warhurst et al26 and DNA extraction with saponin/chelex-100 by Wooden et al27 from 153 dried blood samples on filter paper demonstrated that both methods are sensitive and have high specificity, in addition to being easy and quick to perform. Furthermore, because these methods consist of "in house" protocols, the cost was lower than that of available commercial kits. However, the saponin/chelex-100 method was found to be more sensitive and thus should be the protocol of choice for the DNA-based detection of malaria parasites by nested PCR.
The nested PCR protocol developed by Kimura et al11 for human-infecting Plasmodium species proved to be useful, as it is highly sensitive and specific as well as reproducible and there was no evidence of cross-contamination in the study. Using this protocol, we detected mixed infections that were identified by TS as single infections, which is in accordance with the data from Ebrahimzadeh et al5, Scopel et al21, Singh et al22, Snounou et al24 and Toma et al25.
In mixed infections there is a predominance of one Plasmodium species that generally is the one detected by both TS and nested PCR, whereas the non-dominant species is identified only by PCR-based methods. Moreover, low-level P. malariae parasitemia is especially frequent in mixed infections, probably because of the characteristics of this species and the density-dependant regulation mechanism. Thus, this species could easily be overlooked by microscopy when P. falciparum or P. vivax is the major species. This fact also helps to explain the greater number of mixed infections detected with molecular biology methods compared to conventional microscopic tools27.
PCR-based malaria diagnosis methods are very specific and sensitive but they cannot replace conventional microscopy in routine use9, as they do not allow for the quantification of parasite forms; moreover, they are complex techniques with several steps and high costs. Thus, they can be useful as alternative and complementary tools for the diagnosis of human malaria in developing countries4,13,19,22.
CONCLUSION
The results suggest that there were mixed infections (P. falciparum + P. vivax; P. vivax + P. malariae) in the study area that were not identified by the conventional microscopy method (TS). Furthermore, the two DNA obtainment methods evaluated, washing26 and extraction with saponin/chelex-10027, showed sensitivity and high specificity, as well as lower cost compared to available commercial kits; they were also easy and quick to perform. However, the second method was more sensitive; it provides a simple protocol for DNA extraction from dried blood spots and could be the protocol of choice for the DNA extraction step.
ACKNOWLEDGEMENTS
To Ivana Pimentel and the technicians from the malaria research laboratory of the Instituto Evandro Chagas for their technical assistance.
REFERENCES
1 Alger J, Acosta MC, Lozano C, Velasquez C, Labrada LA. Stained smears as a source of DNA. Mem Inst Oswaldo Cruz. 1996;91(5):589-91.
2 Alves FP, Durlacher RR, Menezes MJ, Krieger H, Silva LH, Camargo EP. High prevalence of asymptomatic Plasmodium vivax and Plasmodium falciparum infections in native Amazonian populations. Am J Trop Med Hyg. 2002;66(6):641-8. [ Links ]
3 Ciceron L, Jaureguiberry G, Gay F, Danis M. Development of a Plasmodium for monitoring efficacy of antimalarial treatment. J Clin Microbiol. 1999;37(1):35-8. [ Links ]
4 Di Santi SM, Kirchgatter K, Brunialti KC, Oliveira AM, Ferreira SR, Boulos M. PCR - based diagnosis to evaluate the performance of malaria reference centers. Rev Inst Med Trop Sao Paulo. 2004 Jul-Aug;46(4):183-7. Doi:10.1590/S0036-46652004000400002 [ Links ]
5 Ebrahimzadeh A, Fouladi B, Fazaeli A. High rate of detection of mixed infections of Plasmodium vivax and Plasmodium falciparum in South-East of Iran, using nested PCR. Parasitol Int. 2007;56(1):61-4. Doi:10.1016/J.Parint.2006.12.001 [ Links ]
6 Edoh D, Steiger S, Genton B, Beck HP. PCR amplifications of DNA from malaria parasites on fixed and stained thick and thin blood films. Trans R Soc Trop Med Hyg. 1997 May-Jun;91:361-3. [ Links ]
7 Farnert A, Arez AP, Correia AT, Bjorkman A, Snounou G, Rosário V. Sampling and storage of blood and detection of malaria parasites by polymerase chain reaction. Trans R Soc Trop Med Hyg. 1999 Jan-Fev;93:50-3. [ Links ]
8 Gilles HM. The malaria parasite. In: Gilles HM, Warrell DA, editors. Essential Malariology. London: Edward Arnold. 1993. p. 12-34.
9 Haghdoost AA, Mazhari S, Bahadini K. Comparing the results of light microscopy with the results of PCR method in the diagnosis of Plasmodium vivax. J Vec Dis. 2006;43:53-7. [ Links ]
10 Imirzalioglu C, Soydan N, Schaller M, Bretzel RG, Chakraborty T, Domann E. Diagnosis of mixed Plasmodium malariae and P. vivax infection in a development aid volunteer by examination of bone-marrow specimens by real-time PCR. J Clin Microbiol. 2006 Jun;44(6):2307-10. Doi: 10.1128/JCM.02687-05 [ Links ]
11 Kimura M, Kaneko O, Liu Q, Zhou M, Kawamoto F, Wataya Y, et al. Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitology Int. 1997;46:91-5.
12 Maeno Y, Nakazawa S, Dao le D, Yamamoto N, Giang ND, Van Hanh T, et al. A dried blood sample on filter paper is suitable for detecting Plasmodium falciparum gametocytes by reverse transcription polymerase chain reaction. Acta Trop. 2008;107(2):121-7. Doi:10.1016/j.actatropica.2008.05.001 [ Links ]
13 McNamara DT, Thomson JM, Kasehagen LJ, Zimmerman PA. Development of a multiplex PCR-ligase detection reaction assay for diagnosis of infection by the four parasite species causing malaria in humans. J Clin Microbiol. 2004 Jun;42(6):2403-10. Doi: 10.1128/JCM.42.6.2403-2410.2004 [ Links ]
14 Ministério da Saúde (BR). Manual de Diagnóstico Laboratorial da Malária. 2. ed. Brasília; 2009. 112 p.
15 Ministério da Saúde (BR), Secretaria de Vigilância em Saúde. Boletim epidemiológico da malária no 02. 2005.
16 Ministério da Saúde (BR). Situação Epidemiológica da Malária no Brasil, ano de 2008. Brasília, 2008. Disponível em: http://portal.saude.gov.br/portal/arquivos/pdf/folder_malaria_2009_web.pdf.
17 Organização Mundial de Saúde. revised July, 2003. Fact sheet no 94. Disponível em: http://www.who.int.dsa.
18 Pan American Health Organization. Regional Strategic Plan for Malaria in the Americas 2006-2010. Washington, D.C: PAHO; 2006. 85 p.
19 Rougemont M, Van Saanen M, Sahli R, Hinrikson HP, Bille J, Jaton K. Detection of Four Plasmodium Species in Blood from Humans by 18S rRNA Gene Subunit-Based and Species-Specific Real-Time PCR Assays. J Clin Microbiol. 2004 Dec;42(12):5636-43. [ Links ]
20 Schindler HC, Montenegro L, Montenegro R, Carvalho AB, Abath FG, Jaureguiberry G. Development and optimization of polymerase chain reaction-based malaria diagnostic methods and their comparison with quantitative buffy coat assay. Am J Trop Med Hyg. 2001 Oct;65(4):355-61. [ Links ]
21 Scopel KK, Fontes CJ, Nunes AC, Horta MF, Braga EM. High prevalence of Plasmodium malariae infections in a Brazilian Amazon endemic area (Apiacás - Mato Grosso State) as detected by polymerase chain reaction. Acta Trop. 2004 Mar;90(1):61-4. Doi:10.1016/J.Actatropica.2003.11.002 [ Links ]
22 Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A Genus - and species-specific nested polymerase chain reaction malaria detection assay for epidemoiologic studies. Am J Trop Med Hyg. 1999 Apr;60(4):687-92. [ Links ]
23 Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993 Apr;58(2):283-92. [ Links ]
24 Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, Rosario VE, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993 Oct;61(2):315-20. [ Links ]
25 Toma H, Kobayashi J, Bouakham V, Arakawa T, Sato Y, Nambanya S, et al. A field study on malaria prevalence in southeastern Laos by polymerase chain reaction assay. Am J Trop Med Hyg. 2001;64(5,6):257-61. [ Links ]
26 Warhurst DC, Awad el Kariem FM, Miles MA. Simplified preparation of malarial blood samples for polymerase chain reaction. Lancet. 1991 Feb;337(8736):303-4. [ Links ]
27 Wooden J, Kyes S, Silbley CH. PCR and strain identification in Plasmodium falciparum. Parasitol Today. 1993 Aug;9(8):303-5.
28 World Health Organization. Malaria diagnosis - new perspectives: report of a joint WHO/USAID informal consultation, 25-27 October 1999. Geneva: WHO; 2000. 1091 p.
 Correspondência / Correspondence / Correspondencia:
Correspondência / Correspondence / Correspondencia:
Giselle Maria Rachid Viana
Laboratório de Pesquisas Básicas em Malária,
Seção de Parasitologia Instituto Evandro Chagas
BR 316, km 7, s/n. 67030-000,
Levilândia. Ananindeua-Pará-Brasil
Phone: 55 91 3214-2148
E-mail:giselleviana@iec.pa.gov.br
Recebido em / Received / Recibido en: 23/7/2009
Aceito em / Accepted / Aceito en: 21/9/2010










 texto em
texto em 
 Curriculum ScienTI
Curriculum ScienTI