Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista Pan-Amazônica de Saúde
versão impressa ISSN 2176-6215versão On-line ISSN 2176-6223
Rev Pan-Amaz Saude v.1 n.3 Ananindeua set. 2010
http://dx.doi.org/10.5123/S2176-62232010000300004
ARTIGO ORIGINAL | ORIGINAL ARTICLE | ARTÍCULO ORIGINAL
Molecular epidemiology of dengue virus serotypes 2 and 3 isolated in Brazil from 1991 to 2008
Epidemiologia molecular dos sorotipos 2 e 3 do vírus dengue, isolados no Brasil de 1991 a 2008
Epidemiología molecular de los serotipos 2 y 3 del virus dengue, aislados en Brasil de 1991 a 2008
Ana Cecília Ribeiro CruzI; Ricardo GallerII; Eliana Vieira Pinto da SilvaI; Mayra de Oliveira e SilvaI; Adriana Ribeiro CarneiroI; Elizabeth Salbé Travassos da RosaI; Helena Baldez VasconcelosI; Eric Luiz Rodrigues de SáIII; Pedro Fernando da Costa VasconcelosI
ISeção de Arbovirologia e Febres Hemorrágicas, Instituto Evandro Chagas/SVS/MS, Ananindeua, Pará, Brasil
IIInstituto de Tecnologia em Imunobiológicos Bio-Manguinhos, Fundação Oswaldo Cruz, Rio de Janeiro, Rio de Janeiro, Brasil
IIIFundação de Medicina Tropical do Tocantins, Araguaina, Tocantins, Brasil
Endereço para correspondência
Correspondence
Dirección para correspondencia
ABSTRACT
The dengue virus (DENV1-4) causes dengue fever and dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS) in tropical and subtropical areas. The aim of this study was to evaluate the circulating genotypes of DENV. This was accomplished by sequencing the PrM and E genes of Brazilian isolates of DENV2 and DENV3 that were obtained between 1991 and 2008 from various geographic regions. Phylogenetic analyses of DENV2 demonstrated that the genotype III (Southeast Asian/American), in spite of several nucleotide and amino acid changes, was the only one that circulated over the past 19 years. Since its introduction in 2000, the DENV3 isolates that have been analyzed have all grouped into genotype III (Indian subcontinent) and there has been no evidence of DENV3 belonging to other genotypes in this study.
Keywords: Dengue Virus; Dengue Hemorrhagic Fever; Flavivirus.
RESUMO
O vírus dengue (DENV1-4) causa a dengue clássica e a febre hemorrágica da dengue / síndrome de choque da dengue (FHD/SCD) em regiões tropicais e subtropicais. O objetivo deste estudo foi avaliar os genótipos circulantes de DENV2 e DENV3 obtidos em distintas regiões geográficas no período de 1991 a 2008. As análises filogenéticas de DENV2 demonstraram que o genótipo III (Sudeste da Ásia/América), apesar das diversas alterações nucleotídicas e de aminoácidos, foi o único a circular durante os últimos 19 anos. Desde a sua introdução no estudo, em 2000, todas as amostras isoladas de DENV3 analisadas foram agrupados no genótipo III (subcontinente indiano). Não foram encontradas evidências de que o DENV3 pertença a outros genótipos investigados.
Palavras-chave: Vírus da Dengue; Febre Hemorrágica da Dengue; Flavivirus.
RESUMEN
El virus dengue (DENV1-4) causa el dengue clásico y la fiebre hemorrágica del dengue / síndrome de choque del dengue (FHD/SCD) en regiones tropicales y subtropicales. El objetivo de este estudio fue el de evaluar los genotipos circulantes de DENV2 y DENV3 obtenidos en distintas regiones geográficas en el período de 1991 a 2008. Los análisis filogenéticos de DENV2 demostraron que el genotipo III (Sudeste de Asia/América), a pesar de las diversas alteraciones nucleotídicas y de aminoácidos, fue el único que circuló durante los últimos 19 años. Desde su introducción en el estudio, en el año 2000, todas las muestras aisladas de DENV3 analizadas, fueron agrupadas en el genotipo III (subcontinente indio). No se encontraron evidencias de que el DENV3 pertenezca a otros genotipos investigados.
Palabras clave: Virus del Dengue; Fiebre Hemorrágica Dengue; Flavivirus.
INTRODUCTION
Dengue fever (DF) and dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS) are major health problems in the tropical and subtropical regions. The increasing number of patients with severe clinical manifestations and the expansion of epidemic areas have led to extensive research on its causative agent, dengue virus (DENV), which is the most widespread and important arthropod-borne virus in terms of morbidity and mortality1,2,3. A global dengue pandemic, which started during World War II has progressively spread worldwide and involves predominantly tropical countries. According to the World Health Organization4, dengue fever results in approximately 5 million hospitalized children and at least 50 thousand deaths are caused by DHF/DSS annually.
The DENV serotypes 1 to 4 (DENV1 to DENV4) belong to the Flavivirus genus of the family Flaviviridae. The genome of flaviviruses is a positive-sense, singlestranded RNA approximately 11 Kb in length, from which a single polyprotein is produced. This polyprotein is proteolytically processed by cellular and viral proteases to generate ten viral proteins. The gene order of the dengue polyprotein is 5'-C-prM(M)-E-NS1-NS2A-NS2B-NS3- NS4A-NS4B-NS5-3'5,6.
Dengue infections can be manifested from asymptomatic to subclinical pictures through DF and DHF/DSS, all of which may be observed during a dengue outbreak. Nonetheless, dengue infections usually result in two well-defined syndromes, DF and DHF. The mechanism by which the DENV-infected host exhibits benign or severe disease remains to be elucidated. The antibody-dependent enhancement (ADE) pathway postulates that circulating antibodies from a primary infection bind to a heterologous virus in a secondary infection facilitating penetration of virus particles into mononuclear cells7,8. Further studies have shown that host immune system mediators, such as cytokines and complement, may have a direct role in the pathogenesis of plasma leakage, a defining feature of DHF9,10,11,12.
In Brazil, DENV2 was first isolated in 1989 from an imported case from Uganda, Africa. Prior to that case, DENV1 and DENV4 caused an epidemic in Boa Vista, Roraima State, Brazil13. In 1986 DENV1 caused epidemics in Rio de Janeiro and additional cities14. In 1990, the first autochthonous DENV2 outbreak occurred in the State of Rio de Janeiro. The introduction of DENV2 increased the number of severe disease cases with the appearance of several cases of DHF/DSS15,16. Furthermore, DENV3 was isolated in São Paulo State from an imported case that occurred in 200017. One year after its introduction into Brazil, DENV3 caused a large dengue outbreak in Rio de Janeiro State and has quickly spread to several other Brazilian States18.
In this study, we have sequenced the C, prM/M and E genes of both DENV2 and DENV3 in 27 Brazilian strains from distinct geographic areas and at different periods of collection between 1991 and 2008 to investigate the molecular epidemiology of these serotypes that are circulating in the Country.
MATERIALS AND METHODS
VIRUS ISOLATION
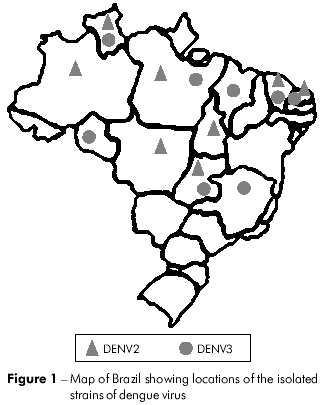
Twenty-seven DENV strains (14 DENV2 and 13 DENV3) were obtained from the virus collection at the Department of Arbovirology and Hemorrhagic Fever in the Instituto Evandro Chagas (IEC). These viruses were isolated from sera collected from patients that were clinically diagnosed as cases of DF, DHF/DSS and encephalitis during the outbreaks that occurred between the period of 1991 to 2008 and from mosquitoes captured in the endemic area (Figure 1). Viruses were grown in cultured Aedes albopictus cells (clone C6/36). Relevant information on each isolate is summarized in table 1.
RNA EXTRACTION AND RT-PCR AMPLIFICATION
Genomic RNA samples were extracted using Trizol reagent ( Invitrogen, USA) according to the manufacturer's instructions. cDNAs were synthesized and amplified with a standard two step RT-PCR assay with specific primers for DENV2 and DENV3 that were des igned to generate over lapping products corresponding to the C/prM/M/E structural genomic region as previously reported. Initially, 0.2-1 µg of viral RNA and 20 µM of antisense primer were heated at 90 oC for 90 s and then placed on ice. RT mix containing 1X RT buffer (200 mM Tris-HCl (pH 8.4), 500 mM KCl), 0.2 mM DNTP (deoxynucleotides) mixture, 100 mM DTT (dithiothreitol), 40 U RNAse inhibitor (Invitrogen, USA) and 200 U Superscript II Reverse transcriptase (Invitrogen, USA) was added to the pre-heated RNA and the volume adjusted to 20 µL. The RT reaction was carried out at 45 oC for 1 h, followed by heating at 94 oC for 10 min. The RT products were used as templates for PCR amplification. Reactions were composed of 20 µM each of the forward and reverse primers, 1X PCR buffer, 1 mM of MgCl2 , 0.2 mM of dNTP mixture, 5 U/µL of Platinum Taq Polymerase (Invitrogen) and the final volume adjusted to 50 µL with RNAse-free water. The samples were placed in a thermal cycler (Perkin Elmer 9600, USA) at 94 oC for 90 s (for initial denaturation) followed by 35 cycles of: 94 oC for 30 s, 55 oC for 30 s, and 72 oC for 120 s and a final extension step at 72 oC for 5 min. RT-PCR products were visualized in a 1.5% agarose gel stained with ethidium bromide (5 µg/mL).
NUCLEOTIDE SEQUENCING AND SEQUENCE ANALYSES
The RT-PCR products were purified using the QlAquick gel extraction kit (Qiagen, Inc., Valencia, CA). Purified cDNA was used as the template for sequencing using the Big Dye Terminator 3.0 kit (Applied Biosystems, Inc., USA) according to the manufacturer's instructions. Sequencing was performed using the ABI Prism 377 (Applied Biosystems) equipment. Nucleotide sequences were analyzed and edited with SeqMan software (DNASTAR, Lasergene software package). Phylogenetic analysis were generated by progressive pairwise multiple sequence alignments using CLUSTAL W (Megalign software; Lasergene software package DNASTAR).
A 2271-base sequence spanning the C-prM-E structural protein genes was used for comparing the 14 isolates described here and 22 previously published sequences of DENV2 retrieved from GenBank (http://www.ncbi.nlm.nih.gov). For DENV3, 1,977 bases from the M and E genes were used to compare our 13 isolates with the 27 previously published sequences of DENV3. The DENV2 sequence (strain Jamaica access number M20558) was used as an outgroup.
Phylogenetic analyses were performed with bootstrap values of 1 thousand pseudoreplicas using the neighbor-joining method (NJ) Kimura-2-parameter19,20,21 (MEGA 2.1 software).
RESULTS
ANALYSIS OF DENV2
We have determined the full nucleotide sequence of the structural genome of 14 DENV2 isolates from patients exhibiting different patterns of disease severity (Table 1). Comparative differences in nucleotide numbers between all viruses studied and the DENV2 Jamaica (Jam83) strain revealed high nucleotide sequence similarity (89.0 to 99.9%) among our isolates. The average similarity between the DENV2 subtypes was 89.5 and 99.8% for the nucleotide and amino acids sequences respectively.
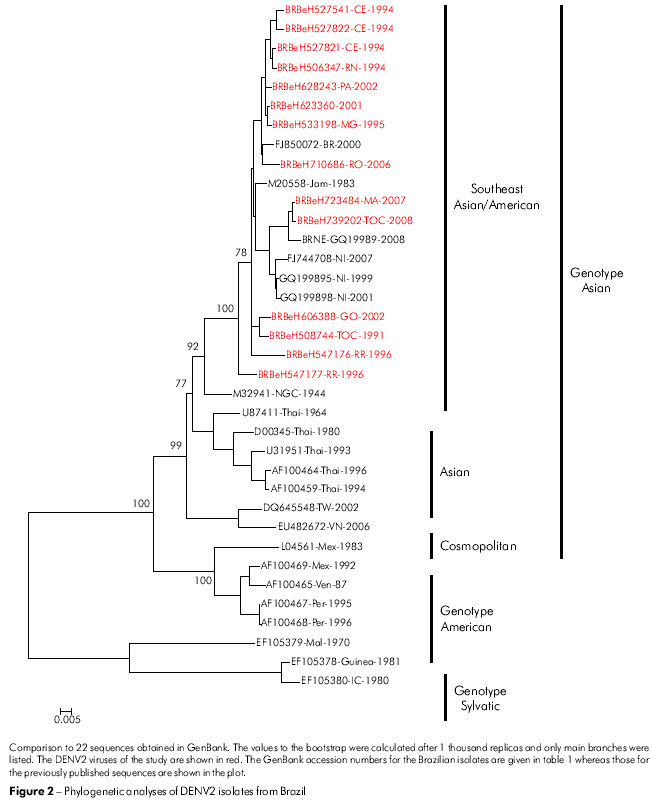
The classification of the genotypes used was proposed in the studies by Rodriguez-Roche22 and Vasilaks and Weaver23 where the sylvatic genotype was used as the outgroup. The phylogenetic tree constructed for the C/prM/M/E nucleotide sequences of our DENV2 isolates grouped all the isolates into the Jamaica genotype (Figure 2; genotype III). In addition, our DENV2 isolates were more closely related to a strain from Brazil (RJ2000; FJ850072) and with the DENV2 Jamaica strain (JAM1983; M20558). The Asian genotype (III) was divided into four different lineages, representing the Southeast Asian/American strains, Asian I, Asian II and Asian III.
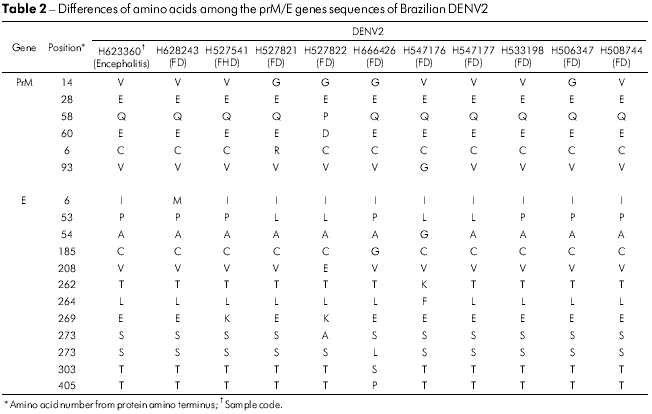
The prM/M/E amino acid sequences of all 14 DENV2 isolates from Brazil showed that these proteins were highly conserved. Only five positions in the prM/M/E genes varied in the five isolates, the most variable being prM amino acid 14, which was a glycine (V → G) in four strains. Isolate BeH527822 accumulated two additional changes at prM/M 58 (Q → P) and 60 (E → D) as shown in table 2. In the E gene, 12 amino acid positions were variable with E53 (P → L) and E269 (E → K) being observed in four and two isolates, respectively. The most variable isolates were BeH527822, BeH666426 and BeH547176 with changes at four positions where the only one that was common to them all was E53 (P → L) (Table 2).
ANALYSIS OF DENV3
Nucleotide sequence homology among the DENV3 strains ranged from 89.7 to 100%. The deduced prM/M and E amino acid sequences for all ten DENV3 isolates showed that their proteins were highly conserved, with a similarity ranging from 93.2 to 100%. The average sequence homology was 94.8% and 96.6% for the nucleotide and amino acid sequences respectively.
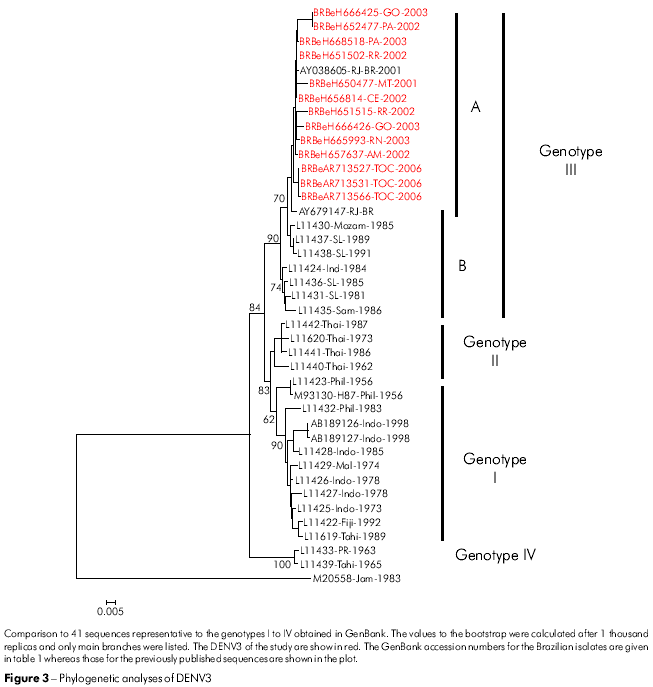
The phylogenetic tree constructed for the DENV3 isolates was based on the alignment of sequences at the E/NS1 region (992-2550 nt), using the criteria for division of the genotypes described by Lanciotti24. All DENV3 isolates were identified as members of genotype III (Indian subcontinent) and clustered separately in two subclades (A and B). Subclade A contains the Latin American strains and is genetically more closely related to subclade B strains isolated in Sri Lanka (L11437, L11438) (Figure 3).
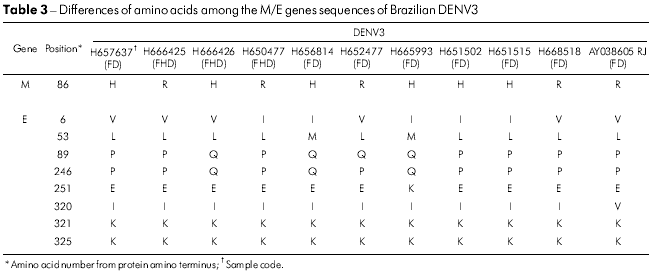
The variability of amino acid sequences for the DENV3 isolates was restricted to one position in the M gene (M86 H → R), whereas six positions were found to vary in the E gene (Table 3). The most variable were E6 (l → V), E89 (Q → P) and E246 (Q → P). Isolates BeH657637 and BeH665993 had accumulated changes in three of the six variable amino acid positions (Table 3).
DISCUSSION
The nucleotide sequence divergence (10.5%) obtained in this study for DENV2 suggests an accumulation of mutations during the period of high circulation of this DENV serotype25. Indeed, DENV2 has circulated in Brazil for almost two decades; however, there has been a surprisingly low incidence of DHF cases when compared to other countries. After introduction of DENV3 in 2002, the number of DHF has increased sharply, suggesting a high virulence of this serotype26. Previous studies on DENV2 from the Americas had shown that the appearance of DHF was related to the introduction of an Asian genotype into the Caribbean Region27. Our data confirms that this DENV2 genotype has been predominant in Brazil throughout the 19 years of its circulation.
According to the classification of Rodriguez-Roche et al22 and Vasilaks and Weaver23, the DENV2 is grouped into three genotypes, with the circulation of genotypes III and IV in the Americas. Genotype III circulated in all countries of the American Region having, as its prototype, the Jamaican strain, which has been associated with DHF cases. The genotype IV (American) has been associated with strains of lower virulence and only associated with DF. It is of interest that this genotype has circulated only in the American subcontinent28. In our study, none of the Brazilian sequences studied were included in the DENV2 genotype IV (American). In fact, all of sequences were included in the genotype III (Figure 2).
The first studies comparing nucleotide sequence of the gene of 12 DENV2 showed no correlation between disease severity and specific nucleotide or amino acid sequence29 . Further analysis of the complete genome of the Southeast Asian DENV2 also failed to identify specific sites that could determine virulence30,31. Leitmeyer et al28 conducted a more consistent analysis of the genetic diversity of DENV2. This study identified an amino acid substitution at position E390 as a primary determinant of severe dengue. In the American genotype, the cases of DF were found to be associated with the presence of an aspartic acid (Asp-D), which may alter interactions with cellular receptors. However, the results obtained with 14 sequences of DENV2 in our study identified an Asn at position E390 in all the sequences studied when the sequences were compared within the 14 strains and with strains of Asian and American genotypes28. The results indicate that the Brazilian DENV2 analyzed have the potential to cause DHF, which is consistent with the data reported with isolates from 32 Venezuela32.
The change of amino acid E390 is located in the carboxyl-terminal domain III (aa 303-395) on the lateral surface, which is believed to contain residues that are involved in tropism and virulence in different flaviviruses33,34,35. In the E gene of 14 Brazilian viruses analyzed, 12 changes were present, and only five amino acids substitutions were significantly associated with a change of character and polarity. Those positions were: E208 (Val/V → Glu/E), 262 (Thr/T → Lys/K), 273 (Ser/S → Leu/L), 274 (Ser/S → Ala/A) and 405 (Thr/T → Pro/P). All amino acid changes are shown in table 2. The changes were present in the ectodomain III or outside of the ectodomain of the protein such as in the residue 405 of sample H666426/Goi4191. Some changes shown in table 2, such as E262 and 264 that are present in H547176/ROR1811 and the viral strain from Venezuela (AF100466), suggest that the virus may have originated from this Country, thus confirming the existence of two modes of entrance of the DENV2 into Brazil.
Over the past two decades, DENV3 has caused epidemics of DHF in Southeast Asia, East Africa and Latin America. The first phylogenetic analysis of the E protein of this dengue serotype resulted in the recognition of four distinct genotypes24. In general, genotypes can be grouped by distinct geographic origin: American, Indian subcontinent, Thailand and Southeast Asian/South Pacific25.
The DENV3 genotype that circulated in the Americas until 1989 had low epidemic potential and was isolated only from DF patients. Between 1980 and 1990, two new genotypes were introduced into the South Pacific and Americas: the Southeast Asian genotype (genotype I) is associated with large epidemics of DHF in Tahiti and Fiji36, and the Indian subcontinent genotype (genotype III) was introduced into Central America in the 1990s in Nicaragua37,38,39,40.
In 1994 DENV3 resurged in the Americas, causing a small outbreak associated with classic DF in Panama. The virus spread toward Northern Central America and reaching Nicaragua and Mexico41,2. Seven years after its introduction into the Americas, DENV3 spread throughout South America reaching Venezuela, Paraguay and Brazil and causing large epidemics42,43,17,44,45.
The 13 Brazilian DENV3 isolates from our study that were collected from different geographical regions, together with those obtained from neighboring countries45,46,40, have all been grouped into genotype III (Indian subcontinent). They are genetically different from the DENV3 strain that previously circulated in the Americas in the 1960s belonging to the genotype V47. It is possible that this accumulated variability is the result of the rapid dispersion and increase in viral replication. This hypothesis is supported by the bootstrap values of around 84% shown in the phylogenic tree based on the neighbor-joining method (Figure 3). The variability of the DENV3 (genotype III) strain may be due to multiple introductions into Brazil, which give rise to subgroups according to origin and year of isolation of the viral strain (Figure 3).
Local transmission of DENV3 was initially detected in Rio de Janeiro State in December 2000. In the summer of 2001, the first autochthonous epidemic occurred in Rio de Janeiro City, with a large number of DHF cases, followed by the spread of this genotype throughout Brazil. All viruses isolated belonged to genotype III, similar to that circulating in Sri Lanka, which is in agreement with previously described studies47,48,49. Our results show, based on the chronology of the virus isolation, that the dispersion of the DENV3 genotype III in Brazil occurred from the Southeast to the Northern region. These results differ from those of Figueiredo et al50, which recorded the circulation of the DENV3 genotype I in the State of Minas Gerais, Southeast to Brazil and Araújo et al51 suggesting the formation of a new genotype (V) that grouped the DENV3 strains from Brazil, China and Japan. The differences in results show that the circulation of new genotypes appeared to be restricted to a specific region in Brazil or to a certain period of time. This raises some important questions: Why have other strains of these genotypes failed to be identified? Does this strain have a potential transmission vector? Therefore, the emergence of other genotypes or novel genotypes should be further investigated. This should include analyzing the complete genome sequences of DENV3 and its epidemiological profile.
CONCLUSION
In summary, our study suggests that the low genetic variability for DENV2 circulating in Brazil may explain the low incidence of severe cases of the disease that have been reported to date. However, the explosive spread of DENV2 genotype III, which has been found to be more variable than DENV3, may have accumulated genetic changes associated with an increase of virulence, immune status and other important factors previously described. This would explain the higher number of severe dengue cases since the introduction of DENV2 and DENV3 into Brazil.
ACKNOWLEDGMENTS
We gratefully acknowledge the valuable support provided by Drs. Márcio Roberto Teixeira Nunes, Ralph Lainson and Elena Caride, Creuza Lima Carvalho and Maria Natividade.
FINANCIAL SUPPORT
This work was partially supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq, Brazil (grants 300460/2005-8 and 501558/2003-9).
REFERENCES
1 Monath TP, Heinz FX. Flaviviruses. In: Fields BN, Knipe DM, Howley PM, editors. Field's Virology. Philadelphia: Lippincott Williams Wilkins; 1996. p. 961-1034.
2 Guzmán MG, Kouri G. Dengue: an update. Lancet Infect Dis. 2002 Jan;2(1):33-42.
3 Kourí G. El dengue, un problema cresciente de salud en las Américas. Rev Panam Salud Publica. 2006;19(3):143-5. [ Links ]
4 World Health Organization. Bulletin of Dengue. 1986
5 Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649-88.
6 Rice CM. Flaviviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams Wilkins; 1996. p. 931-60.
7 Halstead SB. Pathogenesis of dengue: challenges to molecular biology. Science. 1988 Jan;239(4839):476-81. [ Links ]
8 Littaua R, Kurane I, Ennis FA. Human IgG Fc receptor II mediates antibody-dependent enhancement of dengue virus infection. J Immunol. 1990 Apr;144(8):3183-6. [ Links ]
9 Kurane I, Ennis FE. Immunity and immunopathology in dengue virus infections. Semin Immunol. 1992 Apr;4(2):121-7. [ Links ]
10 Rothman AL, Ennis FA. Imunnopathogenesis of dengue haemorrhagic fever. Virology. 1999;257:1-6.
11 Chaturvedi UC, Argwal R, Elbishbishi EA, Mustafá AS. Cytokine cascade in dengue haemorrhagic fever: inplications for pathgenesis. FEMS Immunol Med Microbiol. 2000 Jul;28(3):183-8. [ Links ]
12 Rothman AL. Dengue: defining protective versus pathologic immunity. J Clin Invest. 2004 Apr;113(7):946-51.
13 Travassos da Rosa APA, Travassos da Rosa JFS, Pinheiro FP, Vasconcelos PFC. Arboviroses. In: Leão RNQ, editor. Doenças Infecciosas e Parasitárias. Belém: CEJUP; 1997. p. 207-26.
14 Schatzmayr HG, Nohueira RM, Travassos da Rosa AP. An outbreak of dengue virus at Rio de Janeiro – 1986. Mem Inst Oswaldo Cruz. 1986 Apr-Jun;81(2):245-6. [ Links ]
15 Nogueira RM, Miagostovich MP, Lampe E, Souza RW, Zagne SM, Schatzmayr HG. Dengue epidemic in the stage of Rio de Janeiro, Brazil, 1990-1: co-circulation of dengue 1 and dengue 2 serotypes. Epidemiol Infect. 1993 Aug;111(1):163-70. [ Links ]
16 Zagne SM, Alves VG, Nogueira RM, Miagostovich MP, Lampe E, Tavares W. Dengue haemorrhagic fever in the state of Rio de Janeiro, Brazil: a study of 56 confirmed cases. Trans R Soc Trop Med Hyg. 1994 Nov-Dec;88(6):677-9.
17 Rocco IM, Kavakama BB, Santos CL. First isolation of dengue 3 in Brazil from an imported case. Rev Inst Med Trop Sao Paulo. 2001 Jan-Feb;43(1):55-7.
18 Siqueira Jr JB, Martelli CMT, Coelho GE, Simplício ACR, Hatch DL. Dengue and dengue hemorrhagic fever, Brazil, 1981-2002. Emerg Infect Dis. 2005 Jan;11(1):48-53.
19 Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004 Jun;5(2):150-63. [ Links ]
20 Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406-25. [ Links ]
21 Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J Mol Evol. 1980 Dec;16(2):111-20.
22 Rodriguez-Roche R, Alvarez M, Gritsun T, Halstead S, Kouri G, Gould EA, et al. Virus evolution during a severe dengue epidemic in Cuba, 1997. Virology. 2005;334(2):154-9.
23 Vasilakis N, Weaver SC. History and evolution of human dengue emergence. In: Maramorosch K, Shatkin AS, Murphy FA, editors. Advances in Virus Research. California: Academic Press; 2008. p. 1-76.
24 Lanciotti RS, Lewis JG, Gubler D, Trent DW. Molecular evolution and epidemiology of dengue 3 viruses. J Gen Virol. 1994 Jan;75(1):65-75. [ Links ]
25 Rico-Hesse R. Microevolution and virulence of dengue virus. In: Chambers TJ, Monath TP, editors. The Flaviviruses: structure, replication and evolution. California: Academic Press; 2003. p. 316-41.
26 Secretaria de Vigilância em Saúde. Plano Nacional de Combate a Dengue. Brasília: Ministério da Saúde; 2006.
27 Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, et al. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997 Apr;230(2):224-51.
28 Leitmeyer KC, Vaughn DW, Watts DG, Salas R, Chacon IV, Ramos C, et al. Dengue virus structural differences that correlate with pathogenesis. J Virol. 1999 Jun;73(6):4738-47. [ Links ]
29 Block J, Samuel S, Gibbs AJ, Vitarana UT. Variation of the nucleotide and encoded amino acid sequences of the envelope gene from eight dengue 2 viruses. Arch Virol. 1989;105(1-2):39-53. [ Links ]
30 Mangada MN, IgarashI A. Molecular and in vitro analysis of eigth dengue type 2 viruses isolated from patients exhibiting different disease severities. Virology. 1998 May;244(2):458-66.
31 Pandey BD, Igarashi A. Severity-related molecular differences among nineteen strains of dengue type 2 viruses. Microbiol Immunol. 2000 Feb;44(3):179-88. [ Links ]
32 Uzcategui NY, Camacho D, Comach G, Cuello de Uzcategui R, Holmes EC, Gould EA. Molecular epidemiology of dengue type 2 virus in Venezuela: evidence for in situ virus evolution and recombination. J Gen Virol. 2001 Dec;82(12):2945-53. [ Links ]
33 Hasegawa HM, Yoshida T, Shiosaka S, Kobayashi Y. Mutations in the envelope protein of Japanese encephalitis virus affect entry into cultured cells and virulence in mice. Virology. 1992 Nov;191(1):158-65. [ Links ]
34 Jiang WR, Lowe A, Higgs S, Reid S, Gould EA. Single amino acid codon changes detected in louping ill virus antibody-resistant mutants with reduced neurovirulence. J Gen Virol. 1993 May;74(5):931-5. [ Links ]
35 Mandl CW, Guirakhoo F, Holzmann H, Heinz FX, Kunz C. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J Virol. 1989 Feb;63(2):564-71. [ Links ]
36 Chungue E, Deubel V, Cassar O, Laille M, Martin PMV. Molecular epidemiology of dengue 3 viruses and genetic relatedness among dengue 3 strains isolated from patients with wild or severe form for dengue fever in French Polynesia. J Gen Virol. 1993 Dec;74(12):2765-70.
37 Balmaseda A, Sandoval E, Perez L, Gutierrez CM, Harris E. Application of molecular typing techniques in the 1998 dengue epidemic in Nicaragua. Am J Trop Med Hyg. 1999 Dec;61(6):893-7.
38 Harris E, Roberts TG, Smith L, Selle J, Kramer LD, Valle S, et al. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J Clin Microbiol. 1998 Sep;36(9):2634-9. [ Links ]
39 Harris E, Videa E, Perez L, Sandoval E, Tellez Y, Perez ML, et al. Clinical, epidemiologic, and virologic features of dengue in the 1998 epidemic in Nicaragua. Am J Trop Med Hyg. 2000 Jul-Aug;63(1- 2):5-11. [ Links ]
40 Usuku S, Castillo L, Sugimoto C, Noguchi Y, Yogo Y, Kobayashi N. Phylogenetic analysis of dengue 3 viruses prevalent in Guatemala during 1996-1998. Arch Virol. 2001 Jul;146(7):1381-90. DOI:10.1007/s007050170098 [ Links ]
41 Centers for Disease Control and Prevention. Dengue type 3 infection – Nicaragua and Panama, October-November 1994. MMWR Morb Mortal Wkly Rep. 1995 Jan;44(2):21-4. [ Links ]
42 Halstead SB. Dengue in the Americas and Southeast Asia: do they differ? Rev Panam Salud Publica. 2006 Dec;20(6):407-15. [ Links ]
43 Nogueira RMR, Miagostovich MP, Filippis AM, Pereira MA, Schatzmayr HG. Dengue virus type 3 in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2001 Oct;96(7):925-6. DOI:10.1590/S0074-02762001000700007 [ Links ]
44 Simone TS, Nogueira RM, Araújo ES, Guimarães FR, Santos FB, Schatzmayr HG, et al. Dengue virus surveillance: the co-circulation of DENV1, DENV2 and DENV3 in the State of Rio de Janeiro, Brazil. Trans R Soc Trop Med Hyg. 2004 Sep;98(9):553-62. [ Links ]
45 Aquino VH, Anatriello E, Gonçalves PF, Silva EV, Vasconcelos PF, Vieira DS, et al. Molecular Epidemiology of dengue type 3 virus in Brazil and Paraguay, 2002-2004. Am J Trop Med Hyg. 2006;75(4):710-5. [ Links ]
46 Alvarez M, Rodriguez-Roche R, Bernardo L, Vázquez S, Morier L, Gonzalez D, et al. Dengue Hemorrhagic Fever caused by sequential dengue 1-3 virus infections over a long time interval: Havana epidemic, 2001-2002. Am J Trop Med Hyg. 2006;75(6):1113-7. [ Links ]
47 Uzcategui NY, Comach G, Camacho D, Salcedo M, Cabello de Quintana M, Jimenez M, et al. Molecular epidemiology of dengue virus type 3 in Venezuela. J Gen Virol. 2003 Jun;84(6):1569-75. [ Links ]
48 Messer WB, Gubler DJ, Harris E, Sivananthan K, Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003 Jul;9(7):800-9. [ Links ]
49 Nogueira RM, Schatzmayr HG, Filippis AM, Santos FB, Cunha RV, Coelho JO, et al. Dengue virus type 3, Brazil, 2002. Emerg Infect Dis. 2005 Sep;11(9):1376-81. [ Links ]
50 Figueiredo LB, Cecílio AB, Ferreira GP, Drumond BP, Oliveira JG, Bonjardim CA, et al. Dengue virus 3 genotype 1 associated with dengue fever and dengue hemorrhagic fever, Brazil. Emerg Infect Dis. 2008 Feb;14(2):314-6. [ Links ]
51 Araújo JMG, Bello G, Schatzmayr HG, Santos FB, Nogueira RMR. Dengue virus type 3 in Brazil: a phylogentic perspective. Mem Inst Oswaldo Cruz. 2009;104(3):526-9.
 Correspondência / Correspondence / Correspondencia:
Correspondência / Correspondence / Correspondencia:
Ana Cecília Ribeiro Cruz
Instituto Evandro Chagas,
Seção de Arbovirologia e Febres Hemorrágicas
Rodovia BR 316, km 7, s/nº. Bairro: Levilândia
CEP: 67030-000 Ananindeua-Pará-Brasil
Tel.: 55 (91) 3217-3199 | Fax: 55 (91) 3226-5262
E-mail:anacecilia@iec.pa.gov.br
Recebido em / Received / Recibido en: 31/7/2009
Aceito em / Accepted / Aceito en: 1/2/2010










 texto em
texto em 
 Curriculum ScienTI
Curriculum ScienTI