Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista Pan-Amazônica de Saúde
versão impressa ISSN 2176-6215versão On-line ISSN 2176-6223
Rev Pan-Amaz Saude v.1 n.1 Ananindeua mar. 2010
http://dx.doi.org/10.5123/S2176-62232010000100018
ARTIGO ORIGINAL | ORIGINAL ARTICLE | ARTÍCULO ORIGINAL
Molecular characterization of G1 human rotaviruses detected in children from Belém, Pará, Brazil
Caracterização molecular de rotavírus humanos do tipo G1 detectados em crianças de Belém, Pará, Brasil
Caracterización molecular de rotavirus humano tipo G1 detectado en niños de Belém (Estado de Pará, Brasil)
Luana da Silva SoaresI; Joana D'Arc Pereira MascarenhasI; Yvone Benchimol GabbayI; Rosa Helena Porto GusmãoII; Alexandre da Costa LinharesI
ISeção de Virologia, Instituto Evandro Chagas/SVS/MS, Ananindeua, Pará, Brazil
IICentro de Ciências Biológicas e da Saúde, Universidade do Estado do Pará, Belém, Pará, Brazil
Endereço para correspondência
Correspondence
Dirección para correspondencia
ABSTRACT
Rotavirus is responsible for 40% of gastroenteritis infections worldwide, resulting in 611 thousand deaths annually among infants and young children. The aim of the present study was to perform molecular characterization of strains of the most common circulating rotavirus genotype (G1), which was obtained from children participating in studies previously conducted in Belém, northern Brazil over a 21-year period (1982 to 2003). G1 type rotavirus was detected by polyacrylamide gel electrophoresis, enzyme immunoassay and by reverse transcription polymerase chain reaction for the VP7 and VP4 genes. Of 798 specimens that were found to be positive for rotavirus, 330 (41%) had G1-specificity by EIA using monoclonal antibodies. A total of 148 G1 strains were analyzed by reverse transcription polymerase chain reaction. Electropherotypes and P genotypes characterization of G1 rotavirus occurred at frequencies of 78% and 88%, respectively. Three long electropherotype varieties were identified, with the L1 variety the most frequently found (79%). The G1P[8] combination was the most frequent, responsible for 64% of cases. Mixed infections of G1P[6]+P[8], G1P[4]+P[8], G1P[4]+P[6] and G1P[4]+P[6]+P[8] were found in 11 (7%), 11 (7%), 3 (2%) and 1 (0.6%) samples, respectively. One sample displaying a mixed G1+G4 infection was found. To our knowledge, this is the first study to focus on G1 rotavirus molecular characterization in Brazil. Our findings provide information that will allow a better understanding of the molecular diversity of G1 rotavirus infections in our region.
Keywords: Gastroenteritis; Rotavirus Infections; Genetic Variation.
RESUMO
Os rotavírus são responsáveis por 40% das ocorrências de gastroenterites infantil no mundo, resultando em 611 mil mortes anualmente, e o rotavírus do tipo G1 representa o seu genótipo circulante mais comum. O objetivo do presente artigo foi realizar a caracterização molecular das amostras de rotavírus do tipo G1 obtidas de crianças que participaram de estudos anteriormente conduzidos na Cidade de Belém, norte do Brasil, por um período de 21 anos (1982 a 2003). O rotavírus do tipo G1 foi detectado por meio de eletroforese em gel de poliacrilamida, ensaio imunoenzimático (EIA) e reação em cadeia da polimerase precedida da transcrição reversa para os genes VP7 e VP4. Dos 798 espécimes positivos para rotavírus, 330 (41%) apresentavam especificidade G1 por EIA, usando anticorpos monoclonais. Um total de 148 amostras do tipo G1 foram analisadas por meio da reação em cadeia da polimerase precedida da transcrição reversa. A caracterização dos eletroferotipos e genótipos P dos rotavírus do tipo G1 ocorreu em frequências de 78% e 88%, respectivamente. Três variedades de eletroferotipos longos foram identificados, sendo L1 a predominante (79%). A combinação G1P[8] foi a mais frequente, responsável por 64% dos casos. As infecções mistas G1P[6]+P[8], G1P[4]+P[8], G1P[4]+P[6] e G1P[4]+P[6]+P[8] foram encontradas em 11 (7%), 11 (7%), 3 (2%) e 1 (0,6%) amostras, respectivamente. Uma amostra apresentando infecção mista G1+G4 foi identificada. Ressalte-se que este é o primeiro estudo a abordar a caracterização molecular de rotavírus do tipo G1 no Brasil. Nossos achados permitirão melhor compreensão a respeito da diversidade molecular associada às infecções por rotavírus do tipo G1 em nossa região.
Palavras-chave: Gastroenterites; Infecções por Rotavírus; Variação Genética.
RESUMEN
Los rotavirus son responsables por 40% de los casos de gastroenteritis infantil en el mundo, con más de 611 mil muertes al año, y el rotavirus tipo G1 representa su genotipo mas común en circulación. El objetivo de este trabajo fue realizar la caracterización molecular de las muestras de rotavirus de tipo G1 obtenido en niños que participaron en estudios realizados anteriormente en Belém, ciudad situada en el norte de Brasil, durante un período de 21 años (1982 a 2003). El rotavirus de tipo G1 fue detectado por electroforesis en gel de poliacrilamida, e inmunoensayo enzimático (IEE), y por la reacción en cadena de la polimerasa precedida por transcriptasa inversa para los genes VP7 y VP4. De las 798 especies positivas para rotavirus, 330 (41%) presentaron especificidad G1 por IEE, usando anticuerpos monoclonales. Se analizó mediante la reacción en cadena de la polimerasa precedida de trascripción inversa un total de 148 cepas de tipo G1. La caracterización de genotipos P y electroferotipos de rotavirus de tipo G1 se produjo en las frecuencias de 78% y 88%, respectivamente. Se identificaron tres variedades de electroferotipos largos, siendo el L1 predominante (79%). La combinación G1P [8] fue el más frecuente, representando el 64% de los casos. Las infecciones mixtas G1P[6]+P[8], G1P[4]+P[8], G1P[4]+P[6] y G1P[4]+P[6]+P[8] fueron encontradas en 11 (7%), 11 (7%), 3 (2%) y 1 (0,6%) de las muestras, respectivamente. Se identificó una muestra que presentaba una infección mixta G1+G4. Cabe señalar que este es el primer estudio que analiza la caracterización molecular del rotavirus tipo G1 en Brasil. Nuestros hallazgos harán posible una mejor comprensión de la diversidad molecular asociada a la infección por rotavirus de tipo G1 en nuestra región.
Palabras clave: Gastroenteritis; Infecciones por Rotavirus; Variación Genética.
INTRODUCTION
Group A rotavirus (RV-A) is the most common etiological agent of severe gastroenteritis worldwide. It is responsible for 40% of gastroenteritis infections and results in 611 thousand deaths annually among infants and young children, mostly in the poorest countries. The global magnitude of rotavirus disease is widely recognized; in fact, each child will have an episode of rotavirus gastroenteritis before 5 years of age7,24,25.
Rotavirus belongs to the Reoviridae family and is classified into seven groups (A-G) and four subgroups (I, II, I+II, and non I/II) according to the specificities of epitopes on the inner layer capsid VP6 protein. The rotavirus genome contains 11 segments of double-stranded RNA (dsRNA) inside the core of a tripled-layered capsid. The outer capsid is composed of two structural proteins, VP4 and VP7, which define the P (protease-cleaved protein) and G (glycoprotein) genotypes, respectively. Based on the mobility of the 11 gene segments in polyacrylamide gels, rotaviruses can be 11 identified as long, short or super-short electropherotypes11.
To date, at least 23 G and 30 P types have been identified based on sequence analysis of the VP7 and VP4 genes, respectively; however, few genotypes are known to cause infection in humans5,6,11,31,32. Although theoretically a high number of G/P combinations are possible, epidemiological studies worldwide have documented the circulation of five major types: G1P[8], G2P[4], G3P[8], G4P[8] and G9P[8]6,7,11,19,29.
G1 rotavirus is the most prevalent genotype, and it has been detected in frequencies ranging from 36% to 74% in different regions of the world. G1P[8] strains represent approximately 65% of rotavirus types identified globally29. Castello et al6 have recorded G1P[8] strains in 40% of rotavirus infections in Latin America. Recently, Leite et al19 reported that G1 strains were detected in 43% of cases during the pre-vaccine period in Brazil; by contrast, after vaccine introduction this genotype was reduced to 3% of rotavirus infections due to a large predominance of G2P[4] strains.
Sequence analysis of the VP7 gene of G1 human rotavirus strains in Italy has revealed the existence of at least three VP7 genetic lineages. These antigenic variants might be responsible for the continuous circulation of G1 rotavirus2. Phan et al28 analyzed the VP7 gene of G1 rotavirus strains collected around the world and have suggested a novel nomenclature that includes 11 lineages and 17 sublineages.
The aim of the present study was to perform molecular characterization of G1 rotavirus strains obtained from children participating in various studies previously conducted in Belém, northern Brazil over a 21-year period (1982 to 2003).
MATERIALS AND METHODS
PATIENTS AND CLINICAL SPECIMENS
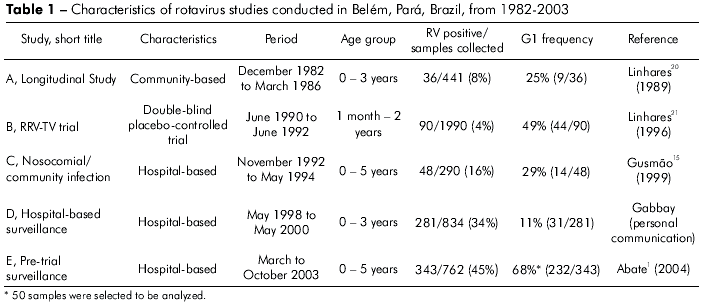
Specimens obtained from five viral gastroenteritis studies conducted in Belém, Brazil, between December 1982 and October 2003, were analyzed for rotavirus detection1,15,20,21. Of 798 specimens that were found to be positive for rotavirus by enzyme immunoassay (EIA), 330 (41%) were G1 typed by EIA using monoclonal antibodies. A total of 148 G1 strains were analyzed by reverse transcription polymerase chain reaction (RT-PCR) to confirm these results. This analysis involved all the G1 strains detected in Studies A, B, C and D, and 20% of the cases from Study E. This study was approved by the Institutional Ethical Review Board from Evandro Chagas Institute, Health Surveillance Secretary.
RNA EXTRACTION
Rotavirus dsRNA was extracted from 10% fecal suspension by using guanidinium isothiocyanate-silica nucleic acid extraction as described previously by Boom et al3.
POLYACRYLAMIDE GEL ELECTROPHORESIS (PAGE)
The RNA profile was analyzed by PAGE with silver staining as described previously16,27.
CHARACTERIZATION OF G AND P ROTAVIRUS GENOTYPES BY REVERSE TRANSCRIPTION POLYMERASE CHAIN REACTION (RT-PCR)
The VP7 genotype was determined by using reverse transcription followed by multiplex PCR as previously published9,14,18. Two different sets of type-specific primers were used for G genotyping: pool A, which contained G1 (9T1-1), G2 (9T1-2), G3 (9T-3P), G4 (9T-4) and G9 (9T-B) specific primers; and pool B which contained primers specific for the G5 (FT5), G6 (DT6), G8 (HT8) and G10 (ET10) genotypes.
Determination of VP4 genotype was performed by RTPCR followed by nested-PCR, as described by Gentsch et al12. Briefly, the full-length VP4 gene was reverse transcribed and a fragment of 876 base pair (bp) was amplified. The primers utilized were specific to P[8] (1T-1), P[4] (2T-1), P[6] (3T-1) and P[9] (4T-1) genotypes.
RESULTS
MOLECULAR EPIDEMIOLOGY OF G1 ROTAVIRUS INFECTION
The highest frequency of G1 rotavirus was found in 2003 during a pediatric hospital-based survey (2003) (68%, 232 of 343), and the lowest was seen during a survey involving hospitalized children (1998 to 2000) (11%, 31 of 281) (Table 1). The median age of the 148 G1-infected children was 13 months (range, 2 month–3 years). Of these, 70 (47%) were <1 year of age, 68 (46%) were 1–2 years of age, and 10 (7%) were >2 years of age.
PAGE OF G1 STRAINS
The RNA profiles were visualized in 116 (78%) of 148 G1 samples tested. Based on migration differences of gene segments 2, 5 and 10, three distinct long electropherotypes were identified (L1, L2 and L3). When the L1 electropherotype is compared with L2, gene segment 2 of L2 migrates more slowly than its corresponding gene in L1. With regards to L3, migration differences for gene segments 5 and 10 were seen with respect to L1. An additional gene segment was found in a sample displaying the L1 pattern (Figure 1). The most frequent electropherotype was L1 (79%, 92 of 116). Study E provided the highest number of samples available for further characterization by PAGE (96%, 48 of 50).
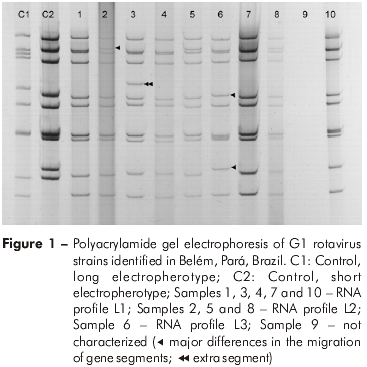
G AND P GENOTYPING
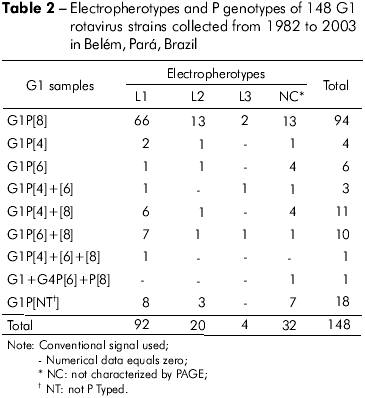
One hundred and forty seven samples bearing G1 specificity and one sample displaying a mixed G1+G4 infection were found. P type could be determined in 130 (88%) samples. P[8] was the most frequent VP4-specificity (64% of cases). In addition, the mixed infections P[6]+P[8], P[4]+P[8], P[4]+P[6] and P[4]+P[6]+P[8] were found in 11 (7%), 11 (7%), 3 (2%) and 1 (0,6%) samples, respectively. Table 2 shows PAGE and RT-PCR (VP4 and VP7 genes) characterization of the 148 samples. The G1P[8] combination displaying the L1 pattern was responsible for 45% of infections.
DISCUSSION
G1 rotavirus still seems to be the most prevalent genotype, representing approximately half of the strains circulating worldwide29. In developed countries, G1 occurs at the highest frequencies (70%-73%). In developing countries, these rates appear to be lower (36%-57%). These differences might be due to the more frequent circulation of unusual strains, as well as the emergence of new types in the poorest countries13,29.
In the present report, the G1 strain was detected in 41% of rotavirus cases over 21 years in studies using different approaches in target populations. Our results are similar to those of a study carried out in Rio de Janeiro, where 50% of samples obtained from children with acute diarrhea were G1 type33. Parra et al26 reported G1 strains in 17% of cases in a study conducted in Paraguay from 1998 to 2000. This difference might be associated with the high prevalence of the G4 type and the detection of the G9 type in the Paraguayan study.
G1 rotavirus rates ranged from 11% to 68% across the analyzed studies. The lowest rates were observed between 1998 and 2000. It is likely that these results are related to the high frequency of untyped strains, as well as to the emergence of G9 during this period. Santos et al30 reported that 79% of all samples analyzed in Salvador were G9. Similar results were also reported in Goiás, where Costa et al8, detected 34% G9 rotavirus infections. These findings suggest that the emergence of G9 in Brazil was followed by wide circulation of strains with this genotype.
It is worth mentioning that all G1 samples displayed long electropherotypes in the present analysis. These RNA profiles showed three different patterns, thus demonstrating a broad diversity of circulating electropherotypes in the region. All varieties were found in specimens from Studies B and D, in which a high frequency of unusual and untyped strains were detected (data not shown). Luz et al22 detected two varieties of long electropherotypes and 19% G1 specimens bearing a short profile with three varieties circulating among children with diarrhea in Maranhão.
The G1P[8] genotype has been found circulating in several countries as the predominant type29. In the present study this genotype was detected in 64% of the samples. Similar results were obtained by Gentsch et al13 and Castello et al6 in studies conducted in Latin America, who found that this strain occurred in 52% and 40% of rotavirus infections, respectively. In Brazil, Carmona et al4 detected G1P[8] in 67% of the specimens collected over eight years in São Paulo.
In the present analysis, one mixed infection involving G types was found (G1+G4). Mascarenhas et al23 analyzed stool samples from a neonate hospitalized for mild/moderate community-acquired diarrhea and noted the occurrence of a G1 plus G4 mixture. Kebaabetswe et al17, in a survey in Botswana involving children with gastroenteritis, detected the mixed infections G1+G2, G1+G8 and G1+G3+G9 in 6%, 22% and 6% of cases, respectively.
It is important to highlight the frequency of mixed infections involving G1 types (17% of cases). Das et al10 reported 30% of cases with mixed infections in an investigation with children hospitalized in India, although other G types were also described. The highest rates of mixed infections are found mostly in developing countries, probably owing to rotavirus gene rearrangements. This might result in the emergence of novel G and P circulating types as well as escape mutants.
To our knowledge, this is the first study focusing on G1 rotavirus molecular characterization in Brazil. Our findings provide information that will allow a better understanding of the molecular diversity of G1 rotavirus infections prior to the advent of RV-vaccine. Further approaches involving nucleotide sequencing might be carried out to assess the circulation of G1 genetic varieties and their possible impact on vaccination strategies.
REFERENCES
1 Abate H, Linhares AC, Venegas G, Vergara R, Lopez P, Jimenez E, et al. Results of a hospital-based study on rotavirus gastroenteritis in Latin American children [abstract]. In: 24th International Congress of Pediatrics (ICP); 2004 Aug 15-20; Cancun, Mexico; 2004. p. 656.
2 Arista S, Giammanco GM, De Grazia S, Ramirez S, Lo Biundo C, Colomba C, et al. Heterogeneity and temporal dynamics of evolution of G1 human rotaviruses in a settled population. J Virol. 2006 Nov;80(21):10724-33. DOI:10.1128/JVI.00340-06 [ Links ]
3 Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheimvan Dillen PM, Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990 Mar;28(3):495-503. [ Links ]
4 Carmona RC, Timenetsky MC, Morillo SG, Richtzenhain LJ. Human rotavirus serotype G9, São Paulo, Brazil, 1996-2003. Emerg Infect Dis. 2006 Jun;12(6):963-8. [ Links ]
5 Castello AA, Argüelles MH, Rota RP, Olthoff A, Jiang B, Glass RI, et al. Molecular epidemiology of group A rotavirus diarrhea among children in Buenos Aires, Argentina, from 1999 to 2003 and emergence of the infrequent genotype G12. J Clin Microbiol. 2006 Jun;44(6):2046-50. DOI: 10.1128/JCM.02436-05 [ Links ]
6 Castello AA, Arvay ML, Glass RI, Gentsch J. Rotavirus strain surveillance in Latin America: a review of the last nine years. Pediatr Infect Dis J. 2004 Oct;23(10 Suppl):S168-72. [ Links ]
7 Centers for Disease Control and Prevention. Rotavirus surveillance—worldwide, 2001-2008. MMWR Morb Mortal Wkly Rep. 2008 Nov;57(46):1255-7. [ Links ]
8 Costa PS, Cardoso DD, Grisi SJ, Silva PA, Fiaccadori F, Souza MB, et al. Rotavirus A infections and reinfections: genotyping and vaccine implications. J Pediatr (Rio J). 2004 Mar-Apr;80(2):119-22. [ Links ]
9 Das BK, Gentsch JR, Cicirello HG, Woods PA, Gupta A, Ramachandran M, et al. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994 Jul;32(7):1820-2. [ Links ]
10 Das S, Varghese V, Chaudhuri S, Barman P, Kojima K, Dutta P, et al. Genetic variability of human rotavirus strains isolated from Eastern and Northern India. J Med Virol. 2004 Jan;72(1):156-61. [ Links ]
11 Estes MK, Kapikian AZ. Rotaviruses. In: Knipe DM, Howley PM, editors. Fields virology. 5th ed. Philadelphia: Lippincott Williams and Wilkins; 2007. p. 1917-74.
12 Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992 Jun;30(6):1365-73. [ Links ]
13 Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, et al. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis. 2005 Sep;192 Suppl 1:S146-59. [ Links ]
14 Gouvea V, Santos N, Timenetsky MC. Identification of bovine and porcine rotavirus G types by PCR. J Clin Microbiol. 1994 May;32(5):1338-40. [ Links ]
15 Gusmão RH, Mascarenhas JD, Gabbay YB, Lins-Lainson Z, Ramos FL, Monteiro TA, et al. Rotavirus subgroups, G serotypes, and electrophoretypes in cases of nosocomial infantile diarrhoea in Belém, Brazil. J Trop Pediatr. 1999 Apr;45(2):81-6. DOI:10.1093/tropej/45.2.81 [ Links ]
16 Herring AJ, Inglis NF, Ojeh CK, Snodgrass DR, Menzies JD. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J Clin Microbiol. 1982 Sep;16(3):473-7. [ Links ]
17 Kebaabetswe LP, Sebunya TK, Matsheka MI, Ndung’u T. Detection and molecular characterisation of group a rotavirus from children in northern Botswana. East Afr Med J. 2005 Apr;82(4):203-8. [ Links ]
18 Leite JP, Alfieri AA, Woods PA, Glass RI, Gentsch JR. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization, and sequence analysis. Arch Virol. 1996;141(12):2365-74. [ Links ]
19 Leite JP, Carvalho-Costa FA, Linhares AC. Group A rotavirus genotypes and the ongoing Brazilian experience: A review. Mem Inst Oswaldo Cruz. 2008 Dec;103(8):745-53. DOI:10.1590/S0074-02762008000800001 [ Links ]
20 Linhares AC, Gabbay YB, Freitas RB, Travassos da Rosa ES, Mascarenhas JD, Loureiro ECB. Longitudinal study of rotavirus infections among children from Belém, Brazil. Epidemiol Infect. 1989 Feb;102(1):129-45. [ Links ]
21 Linhares AC, Gabbay YB, Mascarenhas JD, Freitas RB, Oliveira CS, Bellesi N, et al. Immunogenicity, safety and efficacy of tetravalent rhesus-human, reassortant rotavirus vaccine in Belém, Brazil. Bull World Health Organ. 1996;74(5):491-50. [ Links ]
22 Luz CR, Mascarenhas JD, Gabbay YB, Motta AR, Lima TV, Soares LS, et al. Rotavirus serotypes and electropherotypes identified among hospitalized children in São Luís, Maranhão, Brazil. Rev Inst Med Trop Sao Paulo. 2005 Sep-Oct;47(5):287-93. DOI:10.1590/S0036-46652005000500009 [ Links ]
23 Mascarenhas JD, Leite JP, Lima JC, Heinemann MB, Oliveira DS, Araújo IT, et al. Detection of a neonatal human rotavirus strain with VP4 and NSP4 genes of porcine origin. J Med Microbiol. 2007 Apr;56(Pt 4):524-32. DOI:10.1099/jmm.0.46635-0 [ Links ]
24 Parashar UD, Gibson CJ, Bresse JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006 Feb;12(2):304-6. [ Links ]
25 Parashar UD, Hummelman EG, Bresee JS, Miller MA, Glass RI. Global illness and deaths caused by rotavirus disease in children. Emerg Infect Dis. 2003 May;9(5):565-72. [ Links ]
26 Parra GI, Bok K, Martínez V, Russomando G, Gómez J. Molecular characterization and genetic variation of the VP7 gene of human rotaviruses isolated in Paraguay. J Med Virol. 2005 Dec;77(4):579-86. DOI:10.1002/jmv.20495 [ Links ]
27 Pereira HG, Azeredo RS, Leite JP, Barth OM, Sutmoller F, Farias V, et al. Comparison of polyacrylamide gel electrophoresis (PAGE), immuno-electron microscopy (IEM) and enzyme immunoassay (EIA) for the rapid diagnosis of rotavirus infection in children. Mem Inst Oswaldo Cruz. 1983 Oct-Dec;78(4):483-90. DOI:10.1590/S0074-02761983000400012 [ Links ]
28 Phan TG, Okitsu S, Maneekarn N, Ushijima H. Evidence of intragenic recombination in G1 rotavirus VP7 genes. J Virol. 2007 Sep;81(18):10188-94. DOI:10.1128/JVI.00337-07 [ Links ]
29 Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005 Jan-Feb;15(1):29-56. [ Links ]
30 Santos N, Volotão EM, Soares CC, Campos GS, Sardi SI, Hoshino Y. Predominance of rotavirus genotype G9 during the 1999, 2000, and 2002 seasons among hospitalized children in the city of Salvador, Bahia, Brazil: implications for future vaccine strategies. J Clin Microbiol. 2005 Aug;43(8):4064-9. DOI:10.1128/JCM.43.8.4064-4069.2005 [ Links ]
31 Trojnar E, Otto P, Johne R. The first complete genome sequence of a chicken group A rotavirus indicates independent evolution of mammalian and avian strains. Virology. 2009 Apr;386(2):325-33. DOI:10.1016/j.virol.2009.01.034 [ Links ]
32 Ursu K, Kisfali P, Rigó D, Ivanics E, Erdélyi K, Dán A, et al. Molecular analysis of the VP7 gene of pheasant rotaviruses identifies a new genotype, designated G23. Arch Virol. 2009;154(8):1365-9. DOI:10.1007/s00705-009-0439-0 [ Links ]
33 Volotão EM, Soares CC, Maranhão AG, Rocha LN, Hoshino Y, Santos N. Rotavirus surveillance in the city of Rio de Janeiro-Brazil during 2000-2004: detection of unusual strains with G8P[4] or G10P[9] specificities. J Med Virol. 2006 Feb;78(2):263-72. DOI:10.1002/jmv.20535 [ Links ]
 Correspondência / Correspondence / Correspondencia:
Correspondência / Correspondence / Correspondencia:
Luana da Silva Soares
Instituto Evandro Chagas
Rodovia BR316, km 7, s/no, Levilândia
CEP: 67030-000
Ananindeua-Pará-Brasil
Fax + 00 55 91 32142006
E-mail:luanasoares@iec.pa.gov.br
Recebido em / Received / Recibido en: 31/7/2009
Aceito em / Accepted / Aceito en: 25/9/2009










 texto em
texto em 
 Curriculum ScienTI
Curriculum ScienTI