Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista Pan-Amazônica de Saúde
versão impressa ISSN 2176-6215versão On-line ISSN 2176-6223
Rev Pan-Amaz Saude v.4 n.2 Ananindeua jun. 2013
http://dx.doi.org/10.5123/S2176-62232013000200007
ORIGINAL ARTICLE | ARTIGO ORIGINAL | ARTÍCULO ORIGINAL
Molecular epidemiology of avian rotavirus in fecal samples of broiler chickens in Amazon Region, Brazil, from August 2008 to May 2011
Epidemiologia molecular do rotavirus aviário em fezes de frangos de corte na Região Amazônica, Brasil, de agosto de 2008 a maio de 2011
Epidemiología molecular del rotavirus aviario en heces de pollos de corte en la Región Amazónica, Brasil, de agosto de 2008 a mayo de 2011
René Ribeiro da SilvaI; Delana Andreza Melo BezerraII; Jane Haruko Lima KaianoII; Maria Cristina MannoIII; Darleise de Souza OliveiraII; Fernanda do Espírito Santo SagicaIV; Yvone Benchimol GabbayII; Sílvio Orlan de Castro ChavesI; Artur Luiz da Costa da SilvaV; Amauri Alcindo AlfieriVI; Joana D'Arc Pereira MascarenhasII
ILaboratório Nacional Agropecuário/MAPA, Belém, Pará, Brasil
IISeção de Virologia, Instituto Evandro Chagas/SVS/MS, Ananindeua, Pará, Brasil
IIIInstituto de Saúde e Produção Animal, Universidade Federal Rural da Amazônia, Belém, Pará, Brasil
IVSeção de Meio Ambiente, Instituto Evandro Chagas/SVS/MS, Ananindeua, Pará, Brasil
VInstituto de Ciências Biológicas, Universidade Federal do Pará, Belém, Pará, Brasil
VIDepartamento de Medicina Veterinária Preventiva, Universidade Estadual de Londrina, Londrina, Paraná, Brasil
Correspondence
Endereço para correspondência
Dirección para correspondencia
ABSTRACT
Enteric viruses cause avian diseases that result in economic losses. Avian Rotavirus (AvRV) is the most important virus associated with enteritis in poultry. The main goals of this study were to determine the prevalence of AvRV using molecular tests in broiler chickens (Gallus gallus) from the metropolitan mesoregion of Belém (Pará State, Brazil), to provide epidemiological information, and to compare the rotaviruses detected in this study with reference to strains by phylogenetic analysis. Pooled fecal samples were collected from 37 poultry farms. The samples were tested for the NSP4 gene using reverse transcription polymerase chain reaction (RT-PCR). In total, 35 (41.2%) of the 85 fecal samples were positive for NSP4. There were 19 (51.4%) farms with at least one poultry house positive for AvRV. Considering the distribution of positive samples by age, the chicks aged 31-45 days comprised 18 (51.4%) of the 35 rotavirus-positive samples. Analyzing the data by density population, the houses with more than 9 birds/m2 had 25 (86.2%) positive samples, showing that higher infection rates occurred in higher density houses. To confirm the RT-PCR results and perform phylogenetic analysis, 20 positive samples were selected for sequencing. The rotaviruses detected in our study were clustered in a single group and had 93.5 to 100% sequence identity at the nucleotide level. The most affected age group included broiler chickens older than 15 days. Climatic conditions and high population densities favored the spread of AvRV and supported its uniform maintenance between seasons.
Keywords: Rotavirus; Molecular Epidemiology; Genetics; Virological Analysis.
RESUMO
As viroses entéricas causam doenças em aves que resultam em perdas econômicas. O rotavírus aviário (AvRV) é o vírus mais importante associado à enterite em aves domésticas. Os principais objetivos deste estudo foram determinar a prevalência do AvRV, usando testes moleculares em frangos de corte (Gallus gallus) da mesorregião metropolitana de Belém (Estado do Pará, Brasil), fornecer informações epidemiológicas e comparar os rotavírus detectados com as cepas através da análise filogenética. Amostras fecais foram coletadas em 37 aviários e foram testadas para o gene NSP4 usando a reação em cadeia da polimerase via transcrição reversa (RT-PCR). No total, das 85 amostras fecais, 35 (41,2%) mostram-se positivas para NSP4. Foram encontrados 19 (51,4%) aviários com pelo menos um galpão positivo para AvRV. Considerando a distribuição de amostras positivas por idade, os frangos de 31-45 dias continham 18 (51,4%) das 35 amostras positivas para rotavírus. Analisando os dados pela densidade populacional, os galpões com mais de 9 aves/m2 tiveram 25 (86,2%) amostras positivas, mostrando que índices maiores de infecção ocorreram em galpões de maior densidade. Para confirmar os resultado da RT-PCR e realizar a análise filogenética, 20 amostras positivas foram selecionadas por sequência. Os rotavírus detectados em nosso estudo foram reunidos em um único grupo e tiveram 93,5 a 100% de identidade sequencial no nível de nucleotídeos. O grupo etário mais atingido incluiu frangos de corte com mais de 15 dias. As condições climáticas e a alta densidade populacional favoreceram a disseminação do AvRV e sua manutenção uniforme entre as estações.
Palavras-chave: Rotavirus; Epidemiologia Molecular; Genética; Análise Virológica.
RESUMEN
Las virosis entéricas causan enfermedades en aves que resultan en pérdidas económicas. El rotavirus aviario (AvRV) es el virus más importante asociado a la enteritis en aves domésticas. Los principales objetivos de este estudio fueron de determinar la prevalencia del AvRV usando pruebas moleculares en pollos de corte (Gallus gallus) de la mesorregión metropolitana de Belém (Estado de Pará, Brasil), de suministrar informaciones epidemiológicas y comparar los rotavirus detectados con las cepas a través del análisis filogenético. Fueron recolectadas muestras fecales en 37 pollerías y fueron hechas pruebas para el gen NSP4 usando la reacción en cadena de la polimerasa por transcriptasa reversa (RT-PCR). En total, de las 85 muestras fecales, 35 (41,2%) son positivas para NSP4. Se encontraron 19 (51,4%) pollerías con al menos una jaula positiva para AvRV. Considerando la distribución de muestras positivas por edad, los pollos de 31-45 días contenían 18 (51,4%) de las 35 muestras positivas para rotavirus. Analizando los datos por la densidad poblacional, las jaulas con más de 9 aves/m2 tuvieron 25 (86,2%) muestras positivas, demostrando que mayores índices de infección se hallaron en jaulas con mayor densidad. Para confirmar los resultados de la RT-PCR y realizar el análisis filogenético, se seleccionaron 20 muestras positivas por secuencia. Los rotavirus detectados en nuestro estudio fueron reunidos en un único grupo y tuvieron 93,5 a 100% de identidad secuencial a nivel de nucleótidos. El grupo de edad más afectado incluyó pollos de corte con más de 15 días. Las condiciones climáticas y la elevada densidad poblacional favorecieron la diseminación del AvRV y su manutención uniforme entre las estaciones.
Palabras clave: Rotavirus; Epidemiología Molecular; Genética; Análisis Virológico.
INTRODUCTION
Poultry enteritis is associated with infection by avian Rotavirus (AvRV), Astrovirus, Reovirus, and Coronavirus1,2,3 Molecular methods have been used to determine the prevalence and distribution of enteric viruses in poultry industries due to their high sensitivity and specificity for detecting and identifying viral agents4,5,6.
Rotavirus (RV) is mainly associated with diarrhea in children, other mammals and birds, such as chickens, ducks, pheasants, pigeons and turkeys7,8,9,10,11. The rotaviruses are members of the family Reoviridae and genus Rotavirus, and they have been classified into eight groups/species (A-H)12,13. AvRV A, D, F and G have been isolated from birds14,15,16. Genotypes are classified by differences in the 11 genomic RNA segments17. AvRV may induce enteritis, diarrhea, dehydration, weakness, anorexia, delayed development, poor efficiency in feed conversion and impaired feed utilization, resulting in economic losses to growers18.
AvRV has been detected in several countries, but its potential circulation in Northern Brazil is unknown19,20,21,22. The objectives of this study were to determine the prevalence of the AvRV in the metropolitan mesoregion of Belém (Pará State, Brazil) via molecular testing of broiler chickens and to provide further epidemiological information about this virus. Phylogenetic analysis of the rotaviruses detected in this study was also performed to compare the obtained sequences with reference strains.
MATERIALS AND METHODS
This study was approved by the Ethics Committee on Animal Research of Instituto Evandro Chagas (registration number 0004/2007), and it was conducted in eight counties (Ananindeua, Benevides, Belém, Castanhal, Inhangapi, Santo Antônio do Tauá, Santa Bárbara, and Santa Isabel) in the metropolitan mesoregion of Belém, which is within the northeastern state of Pará, Brazil. This mesoregion is highly involved in poultry production, and the climate is hot and humid (tropical wet), with high annual rainfall. The rainy season occurs between December and May, and the dry season is from June to November, with an average annual temperature of 26o C, 85% relative humidity and 2,870 mm of precipitation23.
Based on official numbers24, there were 180 farms indexed in the mesoregion. From August 2008 to May 2011, fecal samples from poultry litter were collected from 37 commercial chicken flocks belonging to three integrators in eight counties of Pará State. The appropriate number of properties to be included in this study was determined with BioEstat (5.0) software. The farms were chosen based on the availability of integrators, and the number of fecal specimens to be collected was based on the distribution of farms by county. Samples were collected from at least 30% of the poultry houses on each farm, which was visited only once. Nine points were selected at each poultry house, and in each point, a sampling was performed consisting of one pool per house. For convenience, pooled samples are simply referred to as "samples". The ages of these birds ranged from 1 to 45 days, and a total of 85 samples were collected in this period. Three companies agreed to participate and signed a consent form allowing the sampling. No information about the health of the chickens was requested. However, the poultry houses were separated by density conditions according to information obtained in Manno et al25. Therefore, farms were divided into categories representing densities of ≤ 9 birds/m2 and > 9 birds/m2. All samples were stored at -20o C until processed.
A 10% suspension of each sample was prepared in phosphate buffered saline. The suspension was homogenized, and the homogenate was centrifuged at 700 X gfor 10 min. Viral RNA was extracted from 300 µL of stool suspension supernatant with the guanidine isothiocyanate/silica method described by Boom et al26.
All samples were analyzed using a set of specific primers that amplify a 642-bp fragment of the AvRV NSP4 gene27. RT-PCR was performed in two steps. In the first step, 3 µL of extracted RNA and 20 mM of each primer were denatured at 97o C for 5 min in a thermocycler and chilled in an ice bath for 5 min. Reverse transcription was performed in a final volume of 25 µL containing 16.5 µL of nuclease-free H2O, 2.5 µL of 10x buffer (Invitrogen), 1 µl of dNTPs (10 mM, Invitrogen), 0.75 µL of MgCl2 (50 mM, Invitrogen), and 0.25 µL of RT (SuperScript II, 20U, Invitrogen). The reaction was incubated at 42o C for 1 hour. PCR was conducted by adding 25 µL of PCR reagents to the cDNA sample for a final reaction volume of 50 µL. The PCR reagents included 18.5 µL of nuclease-free H2O, 2.5 µL of 10x buffer (Invitrogen), 3 µL of dNTPs (10 mM, Invitrogen), 0.75 µL of MgCl2 (50 mM, Invitrogen), and 0.25 µL of Taq DNA polymerase (2.5 U/µL, Invitrogen), with the following cycling conditions: 94o C for 5 min, 35 cycles of 94o C for 1 min, 50o C for 1 min, and 72o C for 2 min, with a final incubation at 68o C for 7 min. The PCR products were analyzed by gel electrophoresis on a 1.5% agarose gel with TBE buffer, and the gel was stained with SYBR Safe DNA gel stain (Invitrogen). The 642-bp band was visualized with a GEL DOC 1,000 instrument.
At least one positive sample was selected for sequencing from each affected county, except Santa Bárbara, which showed poor PCR product quality. Therefore, 20 samples from 13 different flocks and five different counties were selected. Sequence analysis was performed with conventional RT-PCR products from the initial detection assay. The same samples were purified using the QIAquick PCR Purification Kit (Qiagen), following the protocol provided by the manufacturer.
Purified DNA was sequenced in both directions using a Big Dye Terminator Kit (Applied Biosystems) and an ABI Prism 3130 Genetic Analyzer (Applied Biosystems). The chromatograms were analyzed, and sequences were edited with Bioedit Sequence Alignment Editor software (v. 7.0.9.1). Phylogenetic trees were constructed by MEGA 5.05 software using the neighbor-joining method with a matrix of genetic distances conducted under the Kimura two parameters28 model. Bootstrap analysis with 2,000 replicates assessed the robustness of each node. The reference strains in the GenBank database were compared with the sequenced strains. The analysis included a fragment of 632 nucleotides, which corresponded to nucleotides 35-666 of the NSP4 gene from the group A prototype Ch-02V0002G3 (accession number FJ169862)27.
The data were analyzed using GraphPad Prism software (v. 5.00). Using Fisher's exact test (significance level of p < 0.05), samples from different municipalities, companies, ages, population densities and seasons were tested under the null hypothesis that the presence and absence of the agent were independent of these variables.
The rotavirus nucleotide sequences presented in this article have been submitted to GenBank under accession numbers JN374834 to JN374838 and JX474754 to JX474768.
RESULTS
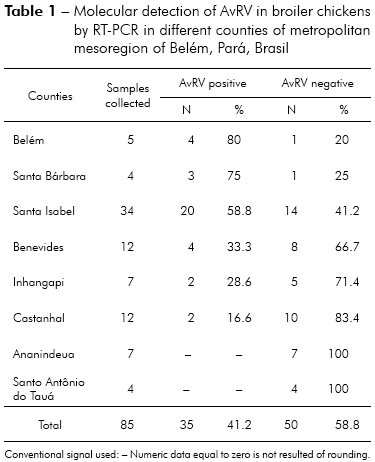
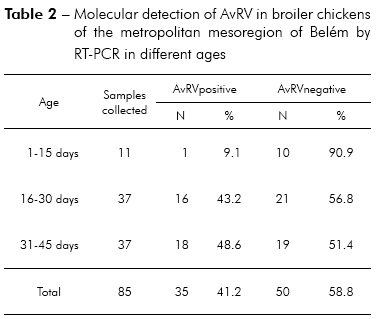
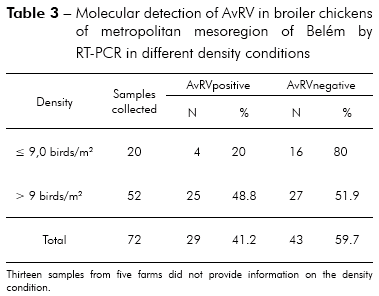
Of the 85 total fecal samples tested, 35 (41.2%) were positive for AvRV. Of the 37 farms studied, 19 (51.4%) had at least one poultry house with AvRV. The virus was present in six (75%) of the eight counties sampled. The highest prevalence was found in Belém, Santa Bárbara and Santa Isabel. Intermediate prevalence was found in Benevides, Inhangapi and Castanhal. There was no virus detected in Ananindeua and Santo Antônio do Tauá (Table 1). Significant differences were observed between Santa Isabel and Castanhal (p ≤ 0.05), Santa Isabel and Ananindeua (p ≤ 0.01), Santa Isabel and Santo Antônio do Tauá (p ≤ 0.05), Castanhal and Belém (p ≤ 0.05), Ananindeua and Belém (p ≤ 0.01), Ananindeua and Santa Bárbara (p ≤ 0.05), Belém and Santo Antônio do Tauá (p ≤ 0.05) and Santa Bárbara and Santo Antônio do Tauá (p ≤ 0.05). All ages and population densities were positive for AvRV. Of the 35 rotavirus-positive samples, 18 (51.4%) were positive in chicks aged 31-45 days old (Table 2). Of the 29 rotavirus-positive samples with available density information, 25 (86.2%) were positive in chicks housed at > 9 birds/m2 (Table 3). There were statistically significant differences between the frequencies observed across age groups (p ≤ 0.01). Significant differences were observed between 1-15 and 16-30 days (p ≤ 0.05) and between 1-15 and 31-45 days (p ≤ 0.05), while no statistically significant differences were observed between 16-30 and 31-45 days (p > 0.05). Significant differences were observed between densities of ≤ 9 and > 9 birds/m2 (p ≤ 0.05). Two (66.7%) of the three integrators had farms with AvRV present. The circulation of the virus across seasons was also observed. There was no statistically significant difference in the frequencies observed between the companies (p > 0.05) and seasons (p > 0.05) studied.
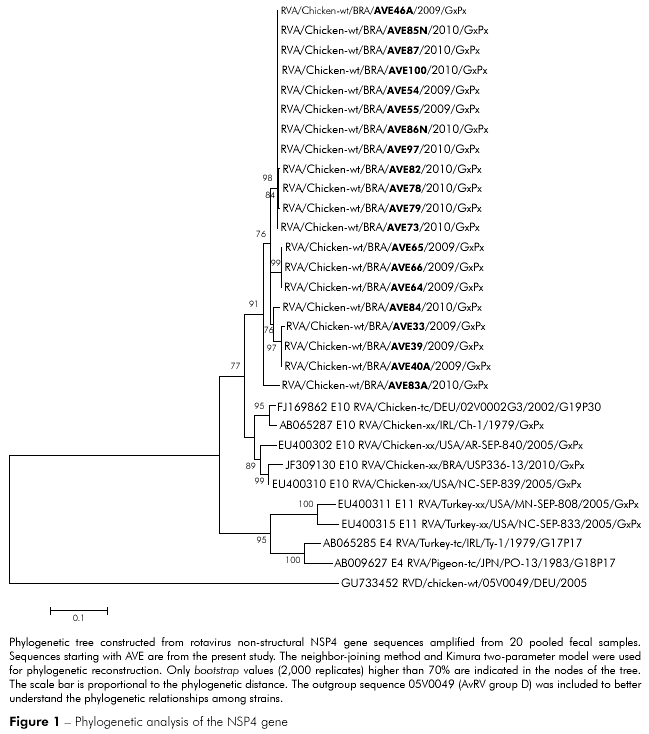
The PCR products from 20 rotavirus-positive samples were sequenced and compared with previously published sequences. Rotaviruses from this study were phylogenetically related to previously reported rotaviruses from Germany (02V0002G3), Ireland (Ch-1), the United States of America (AR/SEP-840; NC/SEP-839) and Brazil (USP336-13), although they were in a separate group. However, AvRV from the metropolitan mesoregion of Belém was unrelated to turkey rotaviruses from Ireland (Ty-1) and the United States of America (NC/SEP-833; MN-SEP-808) and the pigeon rotavirus from Japan (PO-13). Of the 20 rotaviruses isolated in the present study, 19 were phylogenetically related, and only one (AVE83/2010) clustered in a separate clade (Figure 1). The AvRV found in this study had 93.5 to 100% shared sequence identity at the nucleotide level and 93.4 to 100% amino acid identity. A comparison with published chicken virus sequences demonstrated a shared identity of 86.3 to 90.5% at the nucleotide level and 83.2 to 91.8% at the amino acid level. When chicken rotaviruses were compared with previously published turkey rotaviruses, the sequence homology was 67.2 to 73% and 73.1 to 78.3% at the nucleotide and amino acid levels, respectively. Analysis of amino acid positions 1 to 169 of the prototype NSP4 protein 02V0002G3 revealed changes in residues M18I, I43L, R51S, V97I/M, N142T, E146G, E153D and S156N in the predicted proteins of our viral samples.
DISCUSSION
The RT-PCR positivity (41.2%) obtained in this study was lower than that described by Pantin-Jackwood et al2 in healthy commercial turkeys (67.7%) using the same methods and gene with different primers. Subsequently, Jindal et al6 found lower results (30.6%) when testing fecal samples from breeder turkeys by RT-PCR, using the NSP4 gene and the primer used by Pantin-Jackwood et al2. Pantin-Jackwood et al3 detected AvRV in 46.5% of the commercial chickens flocks studied by RT-PCR targeting the NSP4 gene. In this study, we also detected AvRV by RT-PCR in a high number of flocks (51.4%) using primers targeting the NSP4 gene, as described by Trojnar et al27. According to Karim et al29, these differences in results may be due to geoclimatic factors, simultaneous infections, viral immunological aspects, biosecurity and failures in farming practices.
The prevalence of AvRV infection in the studied counties ranged from 0% (Ananindeua and Santo Antônio do Tauá) to 80% (Belém), showing significant variation of the spread of the virus within the studied region (Table 1). Similar findings were reported by Karim et al29, who observed that the prevalence of AvRV infection in chickens showed significant variation across different districts. In this study, the high AvRV prevalence in Belém, Santa Bárbara and Santa Isabel can be explained by the high concentration of farms in these areas. Thus, distances between farms are smaller, and even with the adoption of prophylactic measures by the owner, the risk of dissemination of infectious agents remains high. However, in the areas where AvRV was less prevalent (Santo Antônio do Tauá and Ananindeua), there is a low concentration of farms. Further investigation of the regional density of poultry farms and farm management as risk factors for the spread of infectious agents would clarify the significant differences between the counties studied.
McNulty et al30 reported that AvRV is not normally excreted in the feces of chickens less than 14 days of age, most likely because of passive immunity of maternal origin. Subsequently, McNulty et al31 suggested that the establishment of AvRV in some age groups may occurs by factors such as susceptibility of the younger animals to AvRV, level of passive immunity, infectious dose, differences in farm management and stressful conditions. When studying the presence of rotavirus in birds, Pantin-Jackwood et al3, Islam et al5 and Jindal et al6 found that chicks up to 15 days of age are the most affected age group, with infection frequencies ranging from 37.3 to 45%. Our results identified AvRV by RT-PCR in 48.6% of the samples from chicks 31 to 45 days old. The higher occurrence of rotavirus observed in broiler chickens over 15 days old is not consistent with results in the literature, and the factors described above explain the observed differences. It is important to identify the age group with the highest infection rate to implement effective preventive measures and to avoid infecting future poultry generations.
Greater numbers of positive AvRV tests were observed in animals housed in the highest population densities, with statistically significant differences from the incidence in animals housed in low densities (Table 3). High population densities lead to stressful conditions and elevated levels of virus infections in chicken flocks25,32. The high density of birds most likely facilitated the circulation of AvRV between farms with higher densities (> 9 birds/m2), whereas virus circulation was impaired between farms with lower densities (≤ 9 birds/m2).
The mesoregion studied is the largest producer of broilers in Pará State. Although a number of different companies are developing commercial activities in this area, in some cases, the same company may acquire inputs on the behalf of farms that use different suppliers. In addition, one supplier can supply two or more companies with their products (genetic material, poultry litter, feed, etc.). These factors may explain the lack of significant differences in infection frequencies between the three companies participating in our study.
Seasonality is a characteristic of many diseases and varies by infectious agent. According to Linhares33, the distribution of rotavirus gastroenteritis in Brazil displays a marked seasonal profile, with a higher incidence in temperate regions of the country during the driest months. The Pará State region has a significant incidence of childhood diarrhea by rotavirus during months of low rainfall. In chickens, Barrios et al34 found a uniform seasonal distribution of rotavirus. Karim et al29 detected the highest percentage of rotavirus-positive chicken fecal samples in the summer season. We did not find significant differences between seasons. In this study, the uniform seasonal distribution of rotavirus may be related to the regional climate; there are no large variations in temperature and humidity throughout the seasons.
The genetic similarities between the rotaviruses found in this study and previously published rotaviruses indicate that rotaviruses may cluster within geographic locations3,35. Although rotaviruses from this study were related to previously reported chicken rotaviruses from Germany, Ireland, the United States of America and Brazil, phylogenetic analysis showed that the NSP4 gene sequences from rotaviruses isolated in this study clustered into a separate group, demonstrating the regionality of these samples. However, there were no additional geographical patterns and/or genetic factors suggesting the existence of different genetic groups or variants. Pantin-Jackwood et al2 also studied the NSP4 gene and found four different groups and several variants of AvRV in commercial turkeys. These authors concluded that the identification of numerous viral types and genetic variations within a single flock over time is most likely not a result of divergent adaptation to differing conditions across farms or flocks but is instead due to the chance detection of one variant instead of another.
Schumann et al36 reported the potential for interspecies transmission and reassortment among group A avian rotaviruses. Our results demonstrated that the assortment of chicken rotaviruses occurred in a different group compared with turkey rotaviruses. This result was in agreement with a study by Jindal et al35, which indicates that interspecies transmission of rotaviruses is uncommon.
Research on viral genetic diversity aids the development of new diagnostic techniques and clinical studies, clarifies the distribution of enteric viruses in poultry and elucidates the heterogeneity observed between enteric viruses from different sources. Furthermore, research on genetic diversity can be used to evaluate viruses in humans and the ability of the virus to rearrange27,37.
The NSP4 proteins from mammalian rotaviruses have five functional domains. Only the VP4 binding domain (aa 112-148) and especially the enterotoxin domain (aa 114-135) are conserved in mammalian and bird rotaviruses38. Mori et al39 reported that aa 109-135 from NSP4 was similar among bird viruses and this region has enterotoxin activity in birds. Our results showed that samples from the metropolitan mesoregion of Belém contained viruses with aa 109-135 sequences that were well-conserved compared with the prototype PO-13 (data not shown), suggesting that enterotoxin activity was present in the NSP4 from AvRV detected in our samples. Amino acid analyses are important for evaluating mutations that alter protein functions between strains.
In complete ORF analysis, to consider a strain under study as genotype A, it is necessary that pairwise nucleotide identities between the gene of the strain studied and the strains of a recognized genotype A are over the cut off value (85% for NSP4 gene) of that gene segment17. The samples of the present study had 87.5 to 89.3% sequence identity at the nucleotide level when compared with the prototype E10 (02V0002G3), which leads us to deduce that all samples submitted to nucleotide sequencing of the NSP4 gene in this study could be classified as genotype E10.
CONCLUSION
AvRV circulation was confirmed in the metropolitan mesoregion of Belém, which had areas with high, intermediate and low prevalences of the virus. The most affected group was broiler chickens aged older than 15 days. Population densities > 9 birds/m2 favored the spread of AvRV, while the regional climate supported the uniform maintenance of group A (genotype E10) AvRV between seasons.
ACKNOWLEDGEMENTS
We would like to thank Euzeni Maria de Fátima Menezes for excellent technical assistance.
FINANCIAL SUPPORT
This study was partially supported by a grant from the Science and Technology Secretariat of Pará State, contract no 067/2008 (Sedect/Fapespa/PA), and Instituto Evandro Chagas/SVS/MS, Brazil. Delana Andreza Melo Bezerra and Jane Haruko Lima Kaiano received a Grant fellowship from the Brazilian National Council for the Development of Science and Technology (CNPq).
REFERENCES
1 Tamehiro CY, Alfieri AF, Medici KC, Alfieri AA. Segmented double-stranded genomic RNA viruses in fecal samples from broiler chicken. Braz J Microbiol. 2003;34(4):344-8. Doi:10.1590/S1517-83822003000400013. [Link]
2 Pantin-Jackwood MJ, Spackman E, Day JM, Rives D. Periodic monitoring of commercial turkeys for enteric viruses indicates continuous presence of astrovirus and rotavirus on the farms. Avian Dis. 2007;51(3):674-80. [Link]
3 Pantin-Jackwood MJ, Day JM, Jackwood MW, Spackman E. Enteric viruses detected by molecular methods in commercial chicken and turkey flocks in the United States between 2005 and 2006. Avian Dis. 2008 Jun;52(2):235-44. [Link]
4 Day JM, Spackman E, Pantin-Jackwood M. A multiplex RT-PCR test for the differential identification of turkey astrovirus type 1, turkey astrovirus type 2, chicken astrovirus, avian nephritis virus, and avian rotavirus. Avian Dis. 2007 Sep;51(3):681-4. [Link]
5 Islam MS, Alam MM, Ahmed MU, Saifuzzaman ABM, Kobayashi N, Kayesh MEH, et al. Molecular epidemiologic study on rotavirus infection in human and birds in association with gastroenteritis. Bangladesh J Vet Med. 2009;7(1):233-7. [Link]
6 Jindal N, Patnayak DP, Chander Y, Ziegler AF, Goyal SM. Detection and molecular characterization of enteric viruses in breeder turkeys. Avian Pathol. 2010;39(1):53-61. Doi:10.1080/03079450903490289. [Link]
7 McNulty MS, Curran WL, Todd D, McFerran JB. Detection of viruses in avian faeces by direct electron microscopy. Avian Pathol. 1979 Jul;8(3):239-47. [Link]
8 Takase K, Nonaka F, Sakaguchi M, Yamada S. Cytopathic avian rotavirus isolated from duck faeces in chicken kidney cell cultures. Avian Pathol. 1986;15(4):719-30. [Link]
9 Gough RE, Cox WJ, Devoy J. Isolation and identification of rotavirus from racing pigeons. Vet Rec. 1992 Mar;130(13):273. [Link]
10 Legrottaglie R, Rizzi V, Agrimi P. Isolation and identification of avian rotavirus from pheasant chicks with signs of clinical enteritis. Comp Immunol Microbiol Infect Dis. 1997 Jun;20(3):205-10. [Link]
11 Elschner M, Hotzel H, Reetz J, Diller R, Otto P. Isolation, identification and characterization of group A from a chicken: the inner capsid protein sequence shows only a distant phylogenetic relationship to most other avian group A rotaviruses. Vet Med B Infect Dis Vet Public Health. 2005 Jun;52(5):211-13. Doi:10.1111/j.1439-0450.2005.00848.x. [Link]
12 Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, et al. Family Reoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy. Ninth Report of the ICTV Amsterdam: Elsevier/Academic Press; 2012. p. 541-637.
13 Kindler E, Trojnar E, Heckel G, Otto PH, Johne R. Analysis of rotavirus species diversity and evolution including the newly determined full-length genome sequences of rotavirus F and G. Infect Genet Evol. 2013 Mar;14:58-67. Doi:10.1016/j.meegid.2012.11.015. [Link]
14 Villarreal LYB, Uliana G, Valenzuela C, Chacon JLV, Saindenberg ABS, Sanches AA, et al. Rotavirus detection and isolation from chickens with or without symptoms. Braz J Poult Sci. 2006;8(3):187-91.
15 Bezerra DA, da Silva RR, Kaiano JH, Silvestre RV, de Souza Oliveira D, Linhares AC, et al. Detection of avian group D rotavirus using the polymerase chain reaction for the VP6 gene. J Virol Methods. 2012 Nov;185(2):189-92. Doi: 10.1016/j.jviromet.2012.07.017. [Link]
16 Otto P, Liebler-Tenorio EM, Elschner M, Reetz J, Lohren U, Diller R. Detection of rotaviruses and intestinal lesions in broiler chicks from flocks with runting and stunting syndrome (RSS). Avian Dis. 2006 Sep;50(3):411-8. [Link]
17 Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Bányai K, Estes MK. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol. 2008;153(8):1621-9. Doi:10.1007/s00705-008-0155-1. [Link]
18 Alfieri AF, Tamehiro CY, Alfieri AA. Virus entérico RNA fita dupla, segmentado, em aves: rotavirus, reovirus e picobirnavirus. Semina. 2000;21(1):101-03.
19 Minamoto N, Oki K, Tomita M, Kinjo T, Suzuki Y. Isolation and characterization of rotavirus from feral pigeon in mammalian cell cultures. Epidemiol Infect. 1988 Jun;100(3):481-92. [Link]
20 Polanco G, González M, Manzano L, Cámara J, Puerto M. Rotavirus en animales asintomáticos: detección y clasificación antigénica. Arch Med Vet. 2004;36(1):65-70. Doi:10.4067/S0301-732X2004000100007. [Link]
21 Savita KAL, Malik YPS, Minakshi PG. Detection and characterization of group A and D avian rotaviruses India. Indian J Biotechnol. 2008;7:554-6.
22 Trojnar E, Otto IP, Roth B, Reetz J, Johne R. The genome segments of a group D rotavirus possess group A-like conserved termini but encode group-specific proteins. J Virol. 2010 Oct;84(19):10254-65. Doi:10.1128/JVI.00332-10. [Link]
23 Lima KRS, Alves JAK, Araújo CV, Manno MC, Jesus MLC, Fernandes DL, et al. Inner thermal environment assessment in broiler house with different roofs materials on the great metropolitan region of Belém. Rev Cen Agr. 2009;51:37-50.
24 Agência de Defesa Agropecuária do Estado do Pará. Granjas cadastradas por município da mesoregiáo metropolitana de Belém. Oficio no 225/2006 Belém: ADEPARÁ. 2006.
25 Manno MC, Lima KRS, Aguilar CAL, Souza NSS, Barata ZRP, Viana MAO. Production of ammonia inside poultry houses with environmental changes. Amaz J Agricult Environ Sci. 2011;54:153-8.
26 Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purifications of nucleic acids. J Clin Microbiol. 1990 Mar;28(3):495-503. [Link]
27 Trojnar E, Otto P, Johne R. The first complete genome sequence of a chicken group A rotavirus indicates independent evolution of mammalian and avian strains. Virology. 2009 Apr;386(2):325-33. Doi:10.1016/j.virol.2009.01.034. [Link]
28 Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J Mol Evol. 1980 Dec;16(2):111-20. [Link]
29 Karim MR, Rume FI, Alam MM, Ahmed MU. Molecular epidemiologic study of avian rotavirus prevailing in Bangladesh. Bangl J Vet Med. 2007;5(1-2):43-8. Doi:10.3329/bjvm.v5i1.1308. [Link]
30 McNulty MS, Allan GM, McCracken RM. Experimental infection of chickens with rotaviruses: clinical and virological findings. Avian Pathol. 1983;12(1):45-54. [Link]
31 McNulty MS, Todd D, Allan GM, McFerran JB, Greene JA. Epidemiology of rotavirus infection in broiler chickens: recognition of four serogroups. Arch Virol. 1984;81(1-2):113-21. [Link]
32 Martinez LC, Masachessi G, Carruyo G, Ferreyra LJ, Barril PA, Isa MB, et al. Picobirnavirus causes persistent infection in pigs. Infect Genet Evol. 2010 Oct;10(7):984-8. Doi:10.1016/j.meegid.2010.06.004. [Link]
33 Linhares AC. Rotavirus infection in Brazil: epidemiology and challenges for control. Cad Saude Publica. 2000;16(3):629-46. [Link]
34 Barrios H, Viora S, de Franceschi M, Fliess E. Incidencia de los rotavirus en establecimentos avicolas de producción intensiva. Rev Argent Microbiol. 1991 ene-feb;23(1):15-21. [Link]
35 Jindal N, Patnayak DP, Chander Y, Ziegler AF, Goyal SM. Detection and molecular characterization of enteric viruses from poult enteritis syndrome in turkeys. Poult Sci. 2010 Feb;89(2):217-26. Doi:10.3382/ps.2009-00424. [Link]
36 Schumann T, Hotzel H, Otto P, Johne R. Evidence of interspecies transmission and reassortment among avian group A rotaviruses. Virology. 2009 Apr;386(2):334-43. Doi:10.1016/j.virol.2009.01.040. [Link]
37 Bányai K, Dandár E, Dorsey KM, Mató T, Palya V. The genomic constellation of a novel avian orthoreovirus strain associated with runting-stunting syndrome in broilers. Virus Genes. 2011 Feb;42(1):82-9. Doi:10.1007/s11262-010-0550-z. [Link]
38 Ito H, Sugiyama M, Masubuchi K, Mori Y, Minamoto N. Complete nucleotide sequence of a group A avian rotavirus genome and a comparison with its counterparts of mammalian rotaviruses. Virus Res. 2001 Jun;75(2):123-38. [Link]
39 Mori Y, Borgan MA, Ito N, Sugiyama M, Minamoto N. Sequential analysis of nonstructural protein NSP4 derived from group A avian rotaviruses. Virus Res. 2002. Oct;89(1):145-51. [Link]
 Correspondence / Correspondência / Correspondencia:
Correspondence / Correspondência / Correspondencia:
Joana D'Arc Pereira Mascarenhas
Instituto Evandro Chagas,
Seção de Virologia
Rodovia BR 316, km 7, s/no.
Bairro: Levilândia
CEP: 67030-000
Ananindeua-Pará-Brasil
Tel.: +55 (91) 3214-2016 / Fax: +55 (91) 3214-2005
E-mail: ¡oanamascarenhas@iec.pa.gov.br
Received / Recebido em / Recibido en: 25/2/2013
Accepted / Aceito em / Aceito en: 5/6/2013











 Curriculum ScienTI
Curriculum ScienTI