INTRODUCTION
Mayaro virus (MAYV) and Chikungunya virus (CHIKV) are arboviruses belonging to the Togaviridae family, Alphavirus genus. MAYV was first isolated in 1954 from rural workers in Trinidad and Tobago, and soon thereafter was also isolated in the Guamá river area, in Pará State, Brazil. CHIKV was first isolated in Tanzania, in 1952, during an outbreak attributed to Dengue virus (DENV)1,2,3,4,5.
Outbreaks and epidemics of Mayaro fever were recorded in Brazil, Bolivia, and Peru. In Northern Brazil, there are reports of several outbreaks that occurred in cities of Pará State, such as Belterra (1978), Conceição do Araguaia (1981), and Benevides (1991), and Amazonas State, such as Manaus (2007-2008)6. From 2014 to 2016, 343 human suspected cases of Mayaro fever were reported and distributed in 11 Brazilian States, highlighting Goiás State that presented the highest frequency7. On the other hand, Chikungunya fever presented its first case of autochthony in Americas in 2013, in the Caribbean Region, arriving in Brazil in 2014 and causing tens of thousands of cases8,9. From 2014 to 2017, 505,723 human suspected cases of Chikungunya fever were reported in Brazil9,10,11,12,13.
In humans, MAYV infection is sporadic, mainly affecting people coming from tropical rainforests, where its main vector is the mosquito Haemagogus janthinomys. Regarding CHIKV, a wild and periurban cycle was described in Africa transmitted by Aedes aegypti, and its spread to the Indian Ocean, Asia, and Europe was related to the presence of the vector Aedes albopictus3,4,5,7,14.
The clinical condition of the patient affected by Mayaro fever is a sudden onset characterized by fever reaching 39 °C, headache, myalgia, rash (mainly in the chest, back, arms, and legs), intense pain in the joints of the wrists, ankles, fingers, knees, and elbows, as well as in the lumbar and sacral spine. Similarly, Chikungunya fever is characterized by symmetrical polyarthritis/arthralgia in the wrists, ankles, and elbows, and may regress after 10 days of infection or progress to chronicity4,5,7,14,15,16,17.
The most specific method for the diagnosis of arbovirus infections is reverse transcription polymerase chain reaction (RT-PCR), but virus isolation followed by immunofluorescence (IF) is considered the gold standard method to detect the infectious agent. Attempts of virus isolation can be performed by inoculation in cell cultures of newborn Swiss albino mice (2-3 days old) and eventually also in mosquitoes1,18.
Arthropod cell cultures have been increasingly used, as they have shown sensitivity to virus isolation similar to suckling mice. Among the most used arthropod cell lineages are C6/36 cells from Aedes albopictus, AP-61 cells from Aedes pseudoscutellaris, and TRA-284 cells from Toxorhynchites amboinensis1,16,19. Vertebrate cell cultures are also widely used and include VERO cells from African green monkey kidney (Chlorocebus aethiops), BHK-21 cells from baby hamster kidney (Mesocricetus auratus), and LLC-MK2 cells from rhesus monkey kidney (Macaca mulatta)16,20.
For many arboviruses, little or nothing is known about their eco-epidemiological and molecular characteristics and even their ability to infect various cell lineages currently available. Thus, this work aimed to trace the infection profile of MAYV and CHIKV in four cell lineages, analysing the susceptibility and the permissiveness of BHK-21 and LLC-MK2 cells to alphavirus infections compared to C6/36 and VERO cells, which are more routinely used in specific laboratories. Beside that, studies of virus replication kinetics were performed to follow the progression of virus infection in these cell lineages by assessing cytopathic effect (CPE), virus antigen detection, and progeny RNA load.
MATERIALS AND METHODS
VIRUS SAMPLES
The Brazilian virus isolates used in this study were provided by the arbovirus collection of the Seção de Arbovirologia e Febres Hemorrágicas (SAARB) of the Instituto Evandro Chagas (IEC). MAYV strain (BeAr20290; passage 2; genotype L; genbank accession number KY618127) was isolated from a hematophagous arthropod (Haemagogus sp.) in 1960 and CHIKV (BeH803609; passage 2; Asian genotype; genbank accession number KP164571) was isolated from the blood of a sick human in 2014. The stocks of CHIKV and MAYV used were originated from cell culture fluids and mice brain, respectively, kept in -70 ºC freezer.
CELL CULTURE
The cells used in this study were cultured according to the particularities of each lineage. C6/36 cells (ATCC, USA) were grown at 28 °C in Leibovitz L-15 medium with L-glutamine (Gibco, USA) supplemented with 5% fetal bovine serum (FBS) (Gibco, USA), 3% tryptose phosphate (Himedia, India), 10 mL/L non-essential amino acids (Baktron Microbiology, Brazil), and 1% antibiotic solution (10,000 U/mL penicillin and 10,000 µg/mL streptomycin) (Gibco, USA)16,21. VERO cells (ATCC) were grown at 37 °C and 5% CO2 in medium 199 (Gibco, USA) supplemented with 2.2 g/L sodium bicarbonate (Sigma, USA), 5% FBS and 1% antibiotic solution20,22. BHK-21 cells (ATCC) were grown at 37 °C and 5% CO2 in MEM23 and LLC-MK2 (ATCC) cells were grown at 37 °C and 5% CO2 in DMEM24, both supplemented with 3.7 g/L and 2.2 g/L sodium bicarbonate, respectively, 5% FBS and 1% antibiotic solution.
PREPARATION AND TITRATION OF VIRUS STOCKS
Virus stocks were prepared by inoculating 200 μL of the virus isolates in VERO cells cultured in 25 cm2 culture flasks. The infection was performed by the viral adsorption method. After infection, cells were daily visualized by light microscopy (LM) for the observation of CPE. Cell culture supernatant was obtained from cultures with CPE and fractioned in 500 μL aliquots and stored at -70 °C.
Infectious titers of the virus stocks were determined by plaque assay25 using 10-fold serial dilutions (101 to 107) of each sample prepared in medium 199; 200 μL of each dilution were inoculated into 6-well plates and, after 1 h, 3 mL of semisolid culture medium (1.6% carboxymethyl cellulose in medium 199) supplemented with 2% FBS and 1% antibiotic solution were added. Plates were incubated at 37 °C and 5% CO2 for 48 h and then simultaneously fixed and stained for 3 h in solution containing 1% crystal violet, 30% ethanol, and 20% formalin in phosphate-buffered saline (PBS). Plaques were counted and virus titers were expressed as plaque-forming units (PFU)/mL.
EXPERIMENTAL INFECTION OF CELL LINEAGES
In order to standardize the infecting dose, the multiplicity of infection (MOI) of 1 PFU/cell was used for all cell lineages and 6-well plates were prepared counting the cells according to the virus titer obtained of C6/36, VERO, BHK-21 or LLC-MK2 cells; and then each well plate was infected with MAYV or CHIKV through adsorption of 1 h to perform the analyses of virus replication kinetics at 6, 12, 24, 48, 72, and 96 h post-infection (hpi). A negative control was used for each cell line. All infections were performed according to level three biosafety standards.
PHASE CONTRAST LM (PCLM)
Cells were observed for CPE by pcLM. Phase contrast images were acquired on an Axiovert 25 microscope using 10x/0.25 CP-Achromat objective lens (Zeiss, Germany). Image sets were processed using the ImageJ 1.48 software (NIH, USA).
INDIRECT IMMUNOFLUORESCENCE ASSAY (IIF)
Cells were harvested by scraping and subjected to iIF following a protocol adapted from Gubler et al.26. It was used, as primary antibody, anti-alphavirus polyclonal antibodies produced in mouse, and, as secondary antibodies, anti-mouse antibodies conjugated with fluorescein isothiocyanate (Cappel). iIF images were acquired on a BX51 microscope using a camera with UPlanFL N 20x/0.5 objective lens and WB and U-25ND filters (Olympus, Japan). Image sets were processed using the ImageJ 1.48 software (NIH, USA). A grading amount of positive cells-based score established by visual inspection was used for the expression of fluorescence intensity levels of iIF images as follows: (1+ = 1-25% of positive cells, 2+ = approximately 50% of positive cells, 3+ = approximately 75% of positive cells, and 4+ = approximately 100% of positive cells).
QUANTITATIVE RT-PCR (QRT-PCR)
Cell culture supernatants were harvested by aspiration and subjected to qRT-PCR following the protocols standardized by Celestino Júnior27 and Lanciotti et al.28. Supernatants from infected cell cultures were subjected to RNA extraction using the Maxwell 16 Total RNA Purification kit on the Maxwell 16 instrument (Promega, USA). For the execution of the qRT-PCR technique, the SuperScript III Platinum One-Step qRT-PCR kit with TaqMan probes (Invitrogen, USA) was used on the ViiA 7 Real-Time PCR System (Applied Biosystems, USA). The exponential graphs generated by the amplifications were analyzed on the Prism 6.0 software (GraphPad, USA).
RESULTS
PROGRESSION OF CPE AND TITRATION OF VIRUS STOCK
The CHIKV titer was 5.4 x 107 PFU/mL while the MAYV titer was 3.2 x 108 PFU/mL. In BHK-21 cells (Figure 1A) infected with CHIKV, the manifestation of CPE occurred from 24 hpi; at 48 hpi, the presence of CPE was more evident; and, from 72 hpi, the monolayer was totally destroyed. The same pattern was observed to MAYV in these cells, but the CPE was early detected with just 12 hpi. In virus-infected C6/36 cells (Figure 1B), no CPE was observed. Virus-infected LLC-MK2 cells (Figure 1C) showed relatively later CPE, from 48 hpi in MAYV cultures and 72 hpi in cells infected by CHIKV, which progressed with the destruction of the monolayer at 96 hpi. Virus-infected VERO cells (Figure 1D) showed CPE manifestation from 24 hpi with more prominent effect in MAYV cultures at this time point; and a rapid progression for both viruses towards the destruction of the monolayer in the subsequent time points.
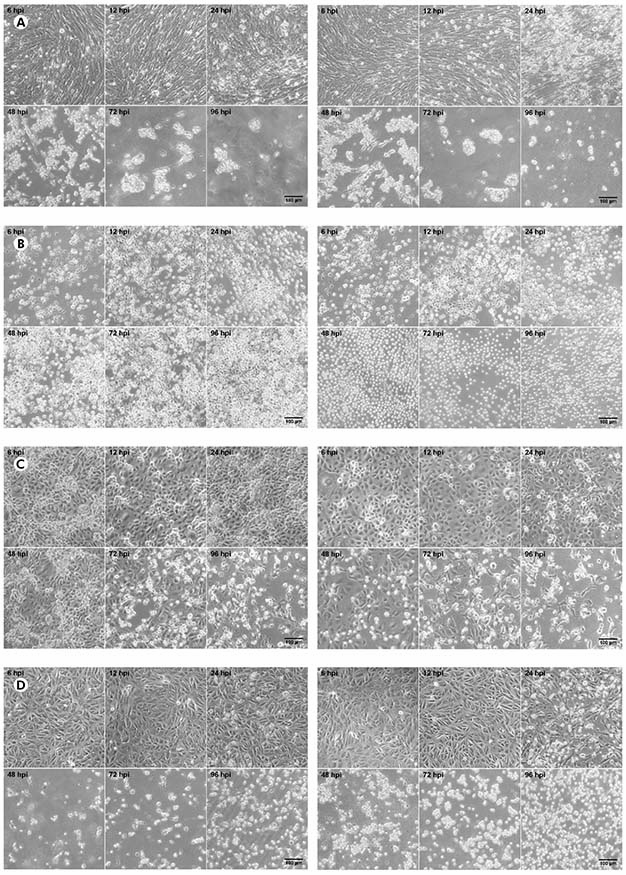
BHK-21 (A), C6/36 (B), LLC-MK2 (C), and VERO (D) cells were infected with CHIKV (left panels) or MAYV (right panels) under the MOI of 1 PFU/cell for the indicated times post-infection, when their transmitted-light images were registered to measure CPE by LM using a phase contrast ring. The images are representative of three visual fields selected randomly for each condition.
Figure 1 - Progression of the CPE in alphavirus-infected cells
In BHK-21, VERO and LLC-MK2 cells infected with both MAYV and CHIKV, the occurrence of lytic type of CPE was observed, causing the rouding of the dead cells, as well as their refraction until complete destruction of the monolayer. No morphological differences were observed between MAYV and CHIKV infected cells. The negative controls of each cell line are presented in the figure 2.
PROGRESSION OF VIRUS ANTIGEN DETECTION
Regarding the iIF of CHIKV-infected BHK-21 cells (Figure 3A, left panels), the positivity of 1+ was observed at 12 hpi and 2+ at 24 hpi; at 48 hpi, the fluorescence progressed to the positivity of 3+; and, at the subsequent times, there was the stabilization of the positivity in 3+. On the other hand, BHK-21 cells infected with MAYV (Figure 3A, right panels) already showed the positivity of 1+ at 6 hpi; at 12 and 24 hpi, there was the positivity of 2+; at 48 hpi, the fluorescence progressed to the positivity of 3+; from 72 hpi on, there was the positivity of 4+.
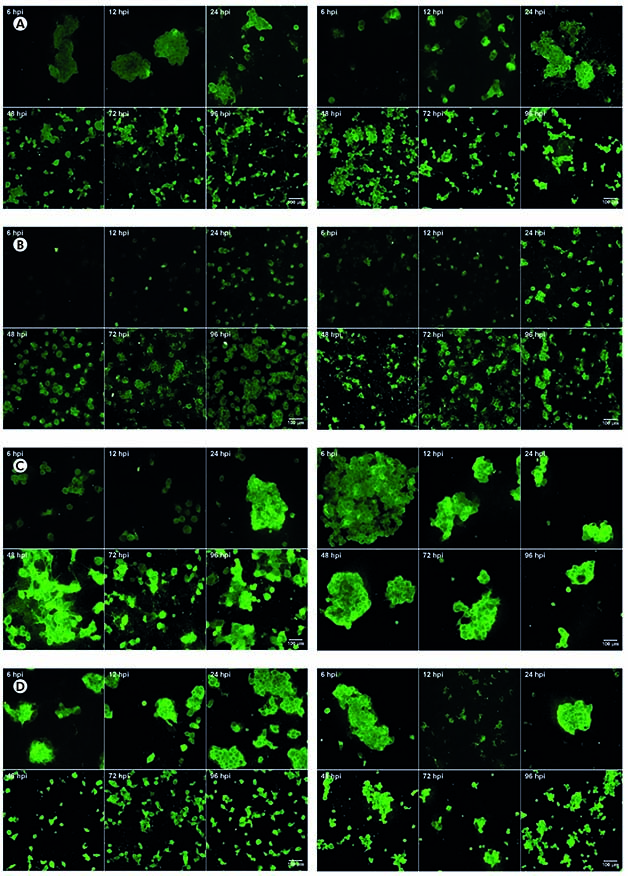
BHK-21 (A), C6/36 (B), LLC-MK2 (C), and VERO (D) cells were infected with CHIKV (left panels) or MAYV (right panels) under the MOI of 1 PFU/cell for the indicated times post-infection, when they were fixed to measure antigen detection by iIF using specific primary antibodies and FITC-labeled secondary antibodies. The images are representative of three visual fields selected randomly for each condition.
Figure 3 - Progression of the virus antigen detection in alphavirus-infected cells
CHIKV-infected C6/36 cells (Figure 3B, left panels) showed just one positive cell at 6 hpi in the iIF, with progression at the subsequent hours: at 12 hpi with the positivity of 2+ and at 24, 48, 72, and 96 hpi with the positivity of 3+. MAYV-infected C6/36 cells (Figure 3B, right panels) showed the positivity of 1+ at 6 hpi and 2+ at 12 hpi, having a progression directly to 3+ at 24 hpi; from 48 hpi on, there was the positivity of 4+.
Concerning CHIKV-infected LLC-MK2 cells (Figure 3C, left panels), the positivity of 1+ was observed at 24 hpi, increasing directly to 3+ from 48 hpi until 96 hpi. In contrast, MAYV-infected LLC-MK2 cells (Figure 3C, right panels) had already showed the positivity of 1+ at 6 hpi and 12 hpi; at 24 hpi and 48 hpi it was observed the positivity of 2+ and 3+, respectively; there was the positivity of 4+ from 72 hpi.
VERO cells infected with CHIKV (Figure 3D, left panels) or MAYV (Figure 3D, right panels) had already showed similar positivities in the iIF assay, with 2+ at 6 hpi and 12 hpi, growing thereafter to 3+ at 24 hpi. Both viruses resulted in the same positivity of 4+ at 48 hpi, with the stabilization of the positivity in 4+ at the subsequent hours.
All cell culture analysis showed higher intensity of fluorescence in those cells infected by MAYV.
The iIF negative controls of each cell line are presented in the figure 4.

BHK-21, C6/36, LLC-MK2, and VERO cells were cultured for 48 h, when they were fixed to measure antigen detection by IF using anti-alphavirus primary antibodies and FITC-labeled secondary antibodies. The images are representative of three visual fields selected randomly for each condition.
Figure 4 - The virus antigen detection in non-infected cells
PROGRESSION OF RNA LOAD IN THE SUPERNATANT
As assessed by qRT-PCR in the supernatant of cell cultures, in CHIKV-infected BHK-21 cells (Figure 5A, black bars), viral RNA was detected at 6 hpi (3.60 x 103 copies/μL), with increase in that value at the subsequent times of 12 hpi (8.68 x 103 copies/μL), 24 hpi (2.60 x 104 copies/μL), 48 hpi (3.31 x 105 copies/μL), and 72 hpi (3.81 x 105 copies/μL). Eventually, there was a drop in viral load at 96 hpi (2.84 x 105 copies/μL). On the other hand, MAYV-infected BHK-21 cells (Figure 5A, gray bars) showed ever-increasing viral load - 4.87 x 102 copies/μL at 6 hpi, 6.85 x 103 copies/μL at 12 hpi, 9.82 x 104 copies/μL at 24 hpi, 6.63 x 105 copies/μL at 48 hpi, 6.87 x 105 copies/μL at 72 hpi, and 1.28 x 106 copies/μL at 96 hpi.
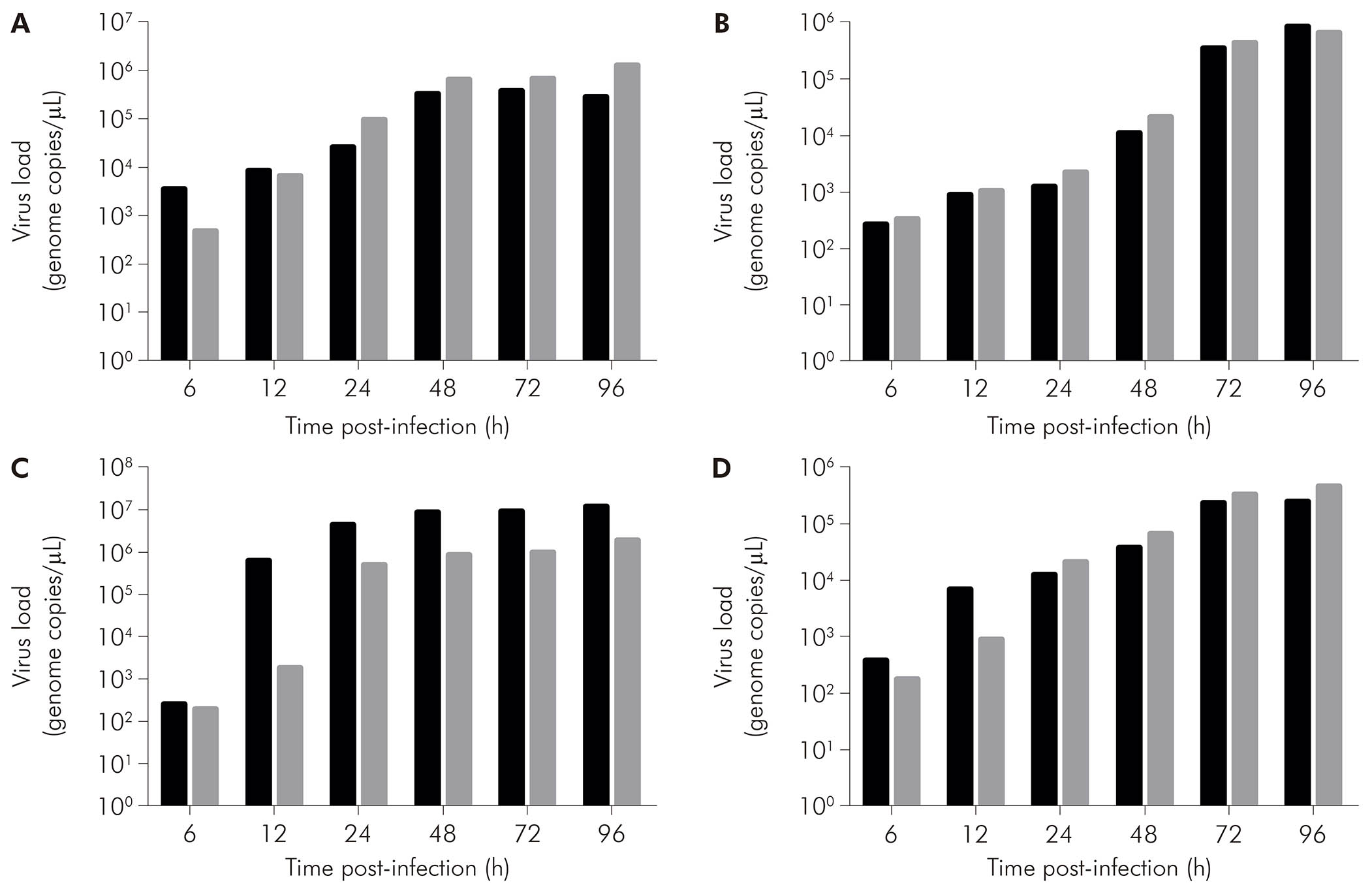
BHK-21 (A), C6/36 (B), LLC-MK2 (C), and VERO (D) cells were infected with CHIKV (black bars) or MAYV (gray bars) under the MOI of 1 PFU/cell for the indicated times post-infection, when their supernatants were harvested to measure RNA load by RT-qPCR using specific primer/probe sets. Data were obtained from three experiments and plotted as mean ± standard deviation for each condition.
Figure 5 - The progression of RNA load in the supernatant of alphavirus-infected cells
CHIKV-infected C6/36 cells (Figure 5B, black bars) had RNA loads of 2.76 x 102 copies/μL at 6 hpi, 9.36 x 102 copies/μL at 12 hpi, 1.27 x 103 copies/μL at 24 hpi, 1.14 x 104 copies/μL at 48 hpi, 3.60 x 105 copies/μL at 72 hpi, and 8.76 x 105 copies/μL at 96 hpi. MAYV-infected C6/36 cells (Figure 5B, gray bars) showed 3.36 x 102 copies/μL at 6 hpi, 1.08 x 103 copies/μL at 12 hpi, 2.25 x 103 copies/μL at 24 hpi, 2.14 x 104 copies/μL at 48 hpi, 4.42 x 105 copies/μL at 72 hpi, and 6.86 x 105 copies/μL at 96 hpi.
CHIKV-infected LLC-MK2 cells (Figure 5C, black bars) showed an increase in RNA load over time: 2.62 x 102 copies/μL at 6 hpi, 6.45 x 105 copies/μL at 12 hpi, 4.54 x 106 copies/μL at 24 hpi, 8.89 x 106 copies/μL at 48 hpi, 9.16 x 106 copies/μL at 72 hpi, and 1.26 x 107 copies/μL at 96 hpi. MAYV-infected LLC-MK2 cells (Figure 5C, gray bars) also showed progressive viral load, exhibiting 2.00 x 102 copies/μL at 6 hpi, 1.84 x 103 copies/μL at 12 hpi, 5.31 x 105 copies/μL at 24 hpi, 8.74 x 105 copies/μL at 48 hpi, 9.97 x 105 copies/μL at 72 hpi, and 1.97 x 106 copies/μL at 96 hpi.
CHIKV-infected VERO cells (Figure 5D, black bars) had RNA loads of 3.98 x 102 copies/μL, 7.00 x 103 copies/μL, 1.28 x 104 copies/μL, 3.98 x 104 copies/μL, 2.48 x 105 copies/μL, and 2.55 x 105 copies/μL at 6 hpi, 12 hpi, 24 hpi, 48 hpi, 72 hpi, and 96 hpi, respectively. MAYV-infected VERO cells (Figure 5D, black bars) had the following RNA loads: 1.82 x 102 copies/μL at 6 hpi, 9.03 x 102 copies/μL at 12 hpi, 2.24 x 104 copies/μL at 24 hpi, 6.91 x 104 copies/μL at 48 hpi, 3.48 x 105 copies/μL at 72 hpi, and 4.88 x 105 copies/μL at 96 hpi.
DISCUSSION
This study aimed to compare the profile of two alphaviruses infection of public health importance in the Brazilian Amazon Region: MAYV, detected for the first time in Brazil in 195529, and CHIKV detected for the first time in Brazil in 201430. The main cell lineages used for the laboratory diagnosis of these arboviruses are C6/36 and VERO, which undoubtedly represent good virus isolation systems. However, few studies have attempted to analyze the susceptibility of other cell lineages, such as BHK-21 and LLC-MK2, in comparison to those previously mentioned, in addition to analyze the onset of detection through iIF and qRT-PCR tests.
This study demonstrates that for both alphaviruses VERO and BHK-21 cells showed earlier CPE, which was already observed at 12 hpi in both cells, when compared to C6/36 and LLC-MK2 cells. C6/36 cells did not show any CPE, as already expected since most arbovirus infections did not lead to death in this cell lineage, and LLC-MK2 cells began to present CPE from 48 hpi. It is noteworthy that the CPE observed in all the cultures that showed it was the lytic type, with complete destruction of the monolayer and large amount of rounded, dead cells.
When comparing MAYV and CHIKV infections, it was observed a more pronounced CPE in cell cultures infected with MAYV (VERO, BHK-21, and LLC-MK2), starting at 12 hpi. In cell cultures infected with CHIKV, the appearance of CPE at 24 hpi was somewhat milder. However, from 48 hpi in general, cell cultures infected by either virus showed a similar pattern of CPE, except for LLC-MK2 cells, which still continued to show a little more advanced CPE when infected with MAYV.
Regarding the results of virus detection by iIF for CHIKV, VERO cells showed the most advanced positivity (at 6 hpi), followed by C6/36 cells, indicating that for this virus these cultures, very used in the laboratory routine, are better for early virus detection by iIF. When the other cell cultures were analyzed, it was observed that, for diagnostic purposes to obtain positivity within a few days, the culture of BHK-21 cells (2+ positivity at 12 hpi) was better than that of LLC-MK2 cells (1+ positivity at 12 hpi).
On the other hand, with regards to MAYV, the iIF assay showed an evolution of the positivity until reaching 4+ in all the cell lineages studied, mentioning that the majority of these cell cultures presented similar pattern of positivity, reaching the positivity of 4+ already at 48 hpi. Thus, it can be suggested that any of the cell lineages studied can be used for diagnostic purposes aimed at early detection of MAYV, and that LLC-MK2 and BHK-21 cells are good alternatives to the lack of C6/36 or VERO cells in the laboratory.
Regarding the RNA loads in cell culture supernatants, by comparing BHK-21 cells infected with MAYV or CHIKV, higher values were obtained for MAYV. This result corroborates the data obtained by iIF, in which MAYV showed the positivity of 4+ and CHIKV showed the positivity of 3+ at the last time points studied. In LLC-MK2 cells, the RNA load was higher with CHIKV than with MAYV, and this was the cell lineage that presented the highest titer among all analyzed cell cultures, pointing out that cells that are not often used in the laboratory routine present good results in virus detection. The qRT-PCR results for LLC-MK2 cells, when compared to the iIF results, may appear to be inconsistent, but it is worth noting that the qRT-PCR assay is more specific than the iIF assay since the first is quantitative and the latter is semi-quantitative and depends on the specificity of the antibodies used.
As for the RNA loads of the C6/36 cell culture, CHIKV reached a higher value at the last analyzed time point (96 hpi); however, at the other times studied, MAYV showed higher values. These results corroborate with those of the iIF assay with regard to higher positivity for MAYV at 48 and 72 h, when compared with CHIKV at the same times. On the other hand, the RNA loads achieved with the VERO cell culture were higher for MAYV at most of the time points of the study (24 to 96 hpi), although the results of the iIF assay indicated similarity between MAYV and CHIKV from the beginning of virus detection at 6 hpi until the last time point tested. However, CHIKV presented higher RNA load values by RT-qPCR in relation to MAYV.
Simon et al.31 reported that the diagnosis of CHIKV is made by RT-PCR and also highlighted the use of C6/36 and VERO cells for the isolation and study of such virus. Given the almost exclusive use of these two cell lineages for arbovirus diagnosis, it is clear the relevance of this study in trying to understand the behavior of alphaviruses in other cell lineages not commonly used in the laboratory routine, such as BHK-21 and mainly LLC-MK2 cells.
In the study by Way et al.32, 20 arboviruses - including nine alphaviruses - were tested in both virus isolation systems (mice and cell culture), and VERO and LLC-MK2 cells had the highest degree of infection when compared to BHK-21 cells. This agrees with the present study showing that the highest RNA load values were obtained on LLC-MK2 cells.
According to Tesh33, the cell lineage most commonly used in the laboratory routine for propagation of arboviruses is C6/36, due to its easy maintenance and growth at 28 °C. Indeed, Figueiredo34 performed the propagation of 23 arboviruses on C6/36 cells, with MAYV being one of the arboviruses studied. No CPE was observed in these cells, and the iIF for alphaviruses only showed positivity after two to three days of infection. These results corroborate the present study, which showed that no CPE was observed for the alphaviruses studied in C6/36 cells; however, differently in this study, virus antigen detection by iIF was possible from 6 hpi, which can be probably explained by the MOI being lower in the Figueiredo's study samples.
Due to the good adaptation of CHIKV to BHK-21 cells, this cell lineage is being currently used to carry out studies of small mutations in virus envelope glycoproteins that facilitate the adaptation of the virus to the arthropod vector35.
Lim and Chu36 performed a replication kinetics of CHIKV in VERO cells infected under the MOI of 10, with the time points of 0, 12, 24, 36, and 48 hpi, and observed CPE from 36 hpi and higher intensification at 48 hpi; in the same study, it was also performed quantification of virus RNA through qRT-PCR at the time points studied, and observed the highest RNA load at 24 hpi, with 5.0 x 108 PFU/mL. In the present study, the replication kinetics with 6, 12, 24, 48, 72, and 96 hpi were performed, with the beginning of CPE in VERO cells infected with CHIKV already at 24 hpi, which corroborates with the study cited36, since the CPE begins to develop from 24 hpi, but at 48 hpi it is already well advanced. However, given the ten-fold lower initial MOI, the RNA load in this study was also lower than that of the Lim and Chu's study, peaking at 96 hpi with 2.55 x 105 copies/μL for CHIKV in VERO cells.
Lee and Chu37 were successful in replicating CHIKV in C6/36 cells to study several proteins that interfere with virus replication in arthropod cells, corroborating the present study by demonstrating that C6/36 cells, although not presenting CPE, provide good results in both iIF and qRT-PCR tests.
Taking together, these findings suggest that VERO, BHK-21 and LLC-MK2 cells are the cells of choice for both CPE monitoring and titration assays by plaque formation. Although C6/36 cells do not demonstrate any CPE for both experimented viruses in question, they have excellent results with early detection, justifying their wide use in laboratory diagnosis. However, this work also shows that other cell lineages less used in the diagnostic routine, such as BHK-21 and especially LLC-MK2, allow excellent RNA loads. In addition, when comparing MAYV with CHIKV, it was usually observed that MAYV achieved higher RNA loads, as well as earlier onset of CPE and iIF detection - this is probably due to the MAYV sample was isolated in 1960, having suffered a greater number of passages than the most recently isolated CHIKV strain.
In addition, works like this one are important to show the best time to harvest infected cells that will serve as virus sources for techniques such as nucleotide sequencing, which requires a large amount of virus RNA; for example, with BHK-21 cell lineage, the highest RNA load achieved for CHIKV was at 72 hpi, and, for MAYV, at 96 hpi. This indicates that there are different replication profiles between viruses in different cell lineages.
Finally, it is well known that many cells used in the laboratory routine are already standardized for the diagnosis of arboviruses, but it is necessary that further studies are carried out with other cell lineages in order to analyze their susceptibility and permissiveness to the increasing number of medically-important arboviruses for the improvement of virological diagnosis.










 text in
text in 
 Curriculum ScienTI
Curriculum ScienTI