Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista Pan-Amazônica de Saúde
Print version ISSN 2176-6215On-line version ISSN 2176-6223
Rev Pan-Amaz Saude vol.10 Ananindeua 2019 Epub Dec 09, 2019
http://dx.doi.org/10.5123/s2176-6223201900096
ORIGINAL ARTICLE
Hepatitis C in the 1980s: case review of former non-A and non-B hepatitis from a hepatology service in the Brazilian Amazon
31 Instituto Evandro Chagas/SVS/MS, Belém, Pará, Brasil
42 Universidade do Estado do Pará, Belém, Pará, Brasil
OBJECTIVE:
To describe the frequency of Hepatitis C virus (HCV) infection in serum samples from patients diagnosed with "non-A and non-B" hepatitis attended at an Amazonian research institute between 1982 and 1988.
MATERIALS AND METHODS:
Descriptive, cross-sectional, and retrospective research including 396 serum samples preserved at -20 °C and tested for anti-HCV IgG antibodies by ELISA. The samples were from patients of both sexes, ranging from 1 month to 85 years old, from Belém and Ananindeua, Pará State, Brazil. For reactive anti-HCV samples, the presence of viral RNA was investigated, and sequencing and genotyping were conducted in those detected.
RESULTS:
Anti-HCV antibodies were detected in 10.9% (43/396) of the sera examined. HCV RNA was detected in 55.8% (24/43) of the reagent sera. For 33.3% (8/24) of these samples, it was possible to distinguish genotypes 1 (75.0%; 6/8) and 3 (25.0%; 2/8), with the confirmation of two subtypes (1b and 3a).
CONCLUSION:
Despite the limitations, due to the long storage time of the samples (three decades), it was possible to detect and characterize HCV in biobank sera, revealing the circulation of two genotypes and two viral subtypes at the time: genotype 1, detected in most genotyped samples and currently exhibiting greater drug resistance than other known genotypes; and genotype 3, less frequent in the study sample, associated with higher virulence. The analysis allowed the identification of HCV strains, favoring future studies to elucidate evolutionary aspects associated with the resistance and virulence of this pathogen.
Keywords: Hepatitis C Virus; Molecular Diagnosis; Genotypic Profile; Epidemiology
INTRODUCTION
Hepatitis C virus (HCV) infection is still a serious public health problem in Brazil and worldwide1. According to the European Association for the Study of the Liver (EASL), 71 million people are chronically infected with HCV around the world, and in the Americas, it is estimated to be 7 million2. Of this number, only 25% were diagnosed2,3. In Brazil between 1.4 and 1.7 million individuals may be infected with HCV4.
Viral hepatitis affects about 325 million people around the world, 95% of those infected are unaware of their disease and less than 1% has access to the treatment3. Hepatitis C is usually asymptomatic, with signs and symptoms occurring mainly in advanced stages of the disease4. Millions of people progress slowly to cirrhosis, decompensated liver disease and hepatocellular carcinoma, making this infection the most frequent cause of liver transplantation in the world1,2,4. Only 10% of patients develop jaundice and less than 20% have more severe nonspecific symptoms, such as anorexia, nausea, vomiting, diarrhea, malaise, abdominal pain, and others5.
Transmission occurs by sharing personal items; syringes and needles for the use of injectable drugs4. The late diagnosis of hepatitis C contributes to the transmission of the disease and its high mortality1,3.
Hepatitis C was first observed in blood donors in the 1980s and characterized as "non-A and non-B" hepatitis (HNANB)6. Initially referred to as "post-transfusion non-A, non-B hepatitis of parenteral transmission", the etiology of hepatitis C was discovered in 19897. The agent of hepatitis C was classified in the family Flaviviridae, genus Hepacivirus and species Hepacivirus C, after the advent of molecular methods8.
Retrospective studies on the frequency of HCV infection are important sources of knowing the natural history of the disease9, but there are few studies in the Brazilian Amazon region about it. This parallel temporal situation also allows the comparison of the epidemiological profile of patients in two different scenarios, before and after the acquisition of diagnostic tests and treatment of that disease. This study aims to describe the prevalence of HCV infection in serum samples from patients diagnosed as HNANB, between 1982 and 1988, from the Legal Amazon.
MATERIALS AND METHODS
This descriptive, cross-sectional, retrospective study used 396 serological samples from the biobank of the Hepatology Section (SAHEP) of the Instituto Evandro Chagas (IEC), the Secretariat of Health Surveillance (SVS), the Ministry of Health (MS), stored at -20 ºC for more than 30 years. These samples were collected from 1982 to 1988 of patients treated with clinical conditions compatible with acute HNANB and, with negative serological markers for hepatitis A and B, from a total of 2,223 consecutive cases treated in the Amazon Region. For the collection of clinical, demographic and laboratory data, the research was carried out in record books and in epidemiological records of the screened cases.
The 396 serum samples were tested for anti-HCV IgG antibodies by Elisa methods of fourth generation kits (DIA.PRO Diagnostic Bioprobes®, San Giovanni, Italy), following the procedures indicated by the manufacturers. The reduced volume of the samples made it impossible to use an automated device in serological diagnosis.
The final result of the samples was given in optical density by reading in spectrophotometer (BIOTEK EL800 Microplate Reader). The cut-off value of this test was calculated by adding the value 0.350 to the average absorbance of the negative controls. The results that exceeded the cut-off value plus 20% were considered reagents. The optical density values below the cut-off value minus 20% were considered non-reactive. Results with optical density up to 20% above or below the cut-off value were characterized as inconclusive.
The samples that presented inconclusive results were submitted to serological test for anti-HCV detection, using Elisa method of third generation kits (ORTHO HCV Version 3.0® ELISA Test System, New Jersey, USA). The interpretations of the research results were carried out by the spectrophotometer (Biotek EL800 Microplate Reader); for the determination of the cut-off values, the constant 0.600 was added to the value of the average absorbance of the negative controls.
Reagent samples for anti-HCV were selected for molecular detection and characterization of HCV. HCV-RNA was researched by RT-qPCR and RT-PCR. RNA extraction was processed in an automated manner (equipment m24sp/m2000rt - Abbott®), with final volume of 60 μL. After extraction, the amplification and detection of HCV-RNA were performed, using the Abbott RealTime HCV kit (Abbott®, USA), according to the manufacturer's instructions. HCV-RNA detection was carried out following TaqMan® systems and using specific Taqman® primers and probes. The Abbott RealTime HCV assay has a linearity of 12 to 100,000,000 IU/mL. The RNA of the samples extracted by automated method (equipment m24sp/m2000rt - Abbott®) was used for the synthesis of cDNA, made with random primers (IDT) under the following thermocycling conditions: 15 min at 25 °C, followed by 1 h to 37 °C and final incubation of 95 °C for 15 min.
The cDNA of the samples was used in RT-PCR assays of the NS5B and 5'UTR regions of HCV partially amplified, subsequent nucleotide sequence and phylogenetic analysis for virus genotyping. The amplification of the NS5B region was developed by heminested RT-PCR assays in order to produce a fragment of 382 base pairs. During the first-round PCR reaction, 5 μL of cDNA of each sample were added to the primers Pr3 (5'TATGAYACCCGCTGYTTTGACTC3') and Pr4 (5'GCNGARTAYCTVGTCATAGTCTC3') and submitted to the thermocycling condition, as described by Morice et al.10 and Sandres-Sauné et al.11. The second-round PCR reaction was performed with primers Pr3 and Pr5 (5' GCTAGTCATAGCCTCCGT3') under cycling conditions10,11. For the 5'UTR region, nested RT-PCR was performed, aiming at amplifying a fragment of 230 base pairs (pb) of the gene encoding the 5'UTR region. 5 μL of cDNA in the PCR reaction and the primers PTC1 (5'CGTTAGTATGAGTGTTGC3') and NCR2 (5'ATACTCGAGGTGCACGGTCTACGACCT3') were used under cycling conditions12. The second PCR was carried out with the primers PTC3 (5'AGTGTCGTGCAGCCTCCAGG3') and NCR4 (5'CACTCTCGAGCACCCTATCAGGCAGT3') under the same cycling conditions as the first PCR.
The detection of amplicons was performed in 1% agarose gel (Ultra Pure Agarose-Invitrogen, Spain), SYBR® Safe DNA Gel Stain (Invitrogen, USA) and molecular weight marker (50 bp DNA Ladder, Invitrogen, USA). The visualization and capture of the images were done with photodetection equipment (TFX-35M GIBCO BRL W Transluminador). Samples with amplification products close to the size expected, 230 bp for the 5'UTR region and 382 bp for the NS5B region were considered positive.
The amplified products were purified with the EXO/SAP-IT kit (GE Healthcare Bio-Sciences, Buckinghamshire, United Kingdom) and subsequently used the sense and antisense sequence with primers Pr3 and Pr5 for the region NS5B and PTC3 and NCR4 for the region 5'UTR, respectively, used with the kit BigDye® Terminator v3.1 Cycle Sequencing (Applied Biosystems, Vilnius, Lithuania) in the automatic sequencer ABI 3500 (Applied Biosystems). All reactions were developed according to the manufacturers' guidelines. The sequences obtained were edited in the Geneious v8.1.3 program and with the sequences of different HCV genotypes and subgenotypes, available in GenBank. Phylogenetic trees were constructed using the Neighbor-Joining method and Kimura 2-parameter model in the MEGA v7 program.
The chi-square test (χ2) of homogeneity was used to compare the proportions of the interest variables (clinical manifestations, sex, age, origin) in two or more populations (infected by HCV or not infected by HCV). The residual analysis showed how the various proportions of the contingency table contribute to the final value of the calculated χ2 (p = 0.05). The BioEstat v5.013 program was used.
In compliance with Resolution No. 466 of December 12, 2012, of the National Health Council/MS14, this project was approved by the Committee for Ethics in Research on Human Beings of the IEC/SVS/MS, under approval opinion no. 1,947,422, on March 3, 2017.
RESULTS
The patients were aged from 1 month to 85 years, with a mean age of 29.9 (± 17.9) and median of 26 years of age, with the majority between 21 and 30 years of age (28.0%). The proportion of men and women was similar, although the frequency for males (55.3%) was bigger. All samples were from patients of the urban area, from the municipalities of Belém and Ananindeua, Metropolitan Region of the state of Pará. The largest number of HNANB patients lived in Marco neighborhood (11.1%). Among the records of patients who had information on occupation, the highest frequency was of students (22.2%). However, there were lack of information in the results for the high frequency of empty fields, e.g. "not informed". The main information on the demographic profile of patients are shown in Table 1.
Table 1 − Distribution of patients diagnosed with HNANB, according to sex, age group, origin and occupation, from 1982 to 1988, in Belém and Ananindeua, Pará State, Brazil
| Variables | N = 396 | % |
|---|---|---|
| Sex | ||
| Male | 219 | 55,3 |
| Female | 177 | 44,7 |
| Age group (years) | ||
| ≤ 10 | 63 | 15,9 |
| 11-20 | 75 | 19,0 |
| 21-30 | 111 | 28,0 |
| 31-40 | 57 | 14,4 |
| 41-50 | 40 | 10,1 |
| 51-60 | 23 | 5,8 |
| ≥ 61 | 27 | 6,8 |
| Origin | ||
| Belém | 349 | 88,1 |
| Ananindeua | 47 | 11,9 |
| Neighborhoods | ||
| Belém | ||
| Marco | 44 | 11,1 |
| Marambaia | 23 | 5,8 |
| Nazaré | 21 | 5,3 |
| Umarizal | 20 | 5,0 |
| Pedreira | 19 | 4,8 |
| Telégrafo | 18 | 4,5 |
| Jurunas | 16 | 4,0 |
| Canudos | 14 | 3,6 |
| Guamá | 14 | 3,6 |
| Sacramenta | 14 | 3,6 |
| São Brás | 14 | 3,6 |
| Icoaraci | 12 | 3,0 |
| Terra Firme | 12 | 3,0 |
| Batista Campos | 11 | 2,7 |
| Coqueiro | 5 | 1,3 |
| Cremação | 9 | 2,3 |
| Val-de-Cans | 5 | 1,3 |
| Campina | 4 | 1,0 |
| Souza | 4 | 1,0 |
| Other neighborhoods | 19 | 4,8 |
| Not informed | 51 | 12,8 |
| Ananindeua | ||
| Cidade Nova | 16 | 4,0 |
| Coqueiro | 6 | 1,5 |
| Guanabara | 5 | 1,3 |
| Other neighborhoods | 14 | 3,6 |
| Not informed | 6 | 1,5 |
| Occupation | ||
| Student | 88 | 22,2 |
| Housekeeper | 38 | 9,6 |
| Minor without occupation | 33 | 8,3 |
| Homemaker | 24 | 6,1 |
| Merchant | 14 | 3,5 |
| Teacher | 9 | 2,3 |
| Public employee | 7 | 1,8 |
| Driver | 7 | 1,8 |
| Construction worker | 7 | 1,8 |
| Retired | 6 | 1,5 |
| Office assistant | 4 | 1,0 |
| Other occupations | 82 | 20,7 |
| Not informed | 77 | 19,4 |
N: Sample number; %: Frequency.
The clinical manifestations reported by the patients, who sought the SAHEP/IEC in the 1980s and who received the diagnosis by HNANB, were collected from 322 of the 396 completed and available forms, highlighting: choluria (180/322; 55.9%), jaundice (161/322; 50.0%), fever (148/322; 46.0%), vomiting (106/322; 32.9%), myalgia (93/322; 28.9%), malaise (90/322; 27.9%), anorexia (76/322; 23.6%), nausea (76/322; 23.6%) diarrhea (41/322; 12.7%).
The individuals who tested positive for anti-HCV living in Belém and were mostly from Marco neighborhood (6/38; 15.8%). Figure 1 highlights neighborhoods with two or more cases of anti-HCV reagents (23/38; 60.5%).
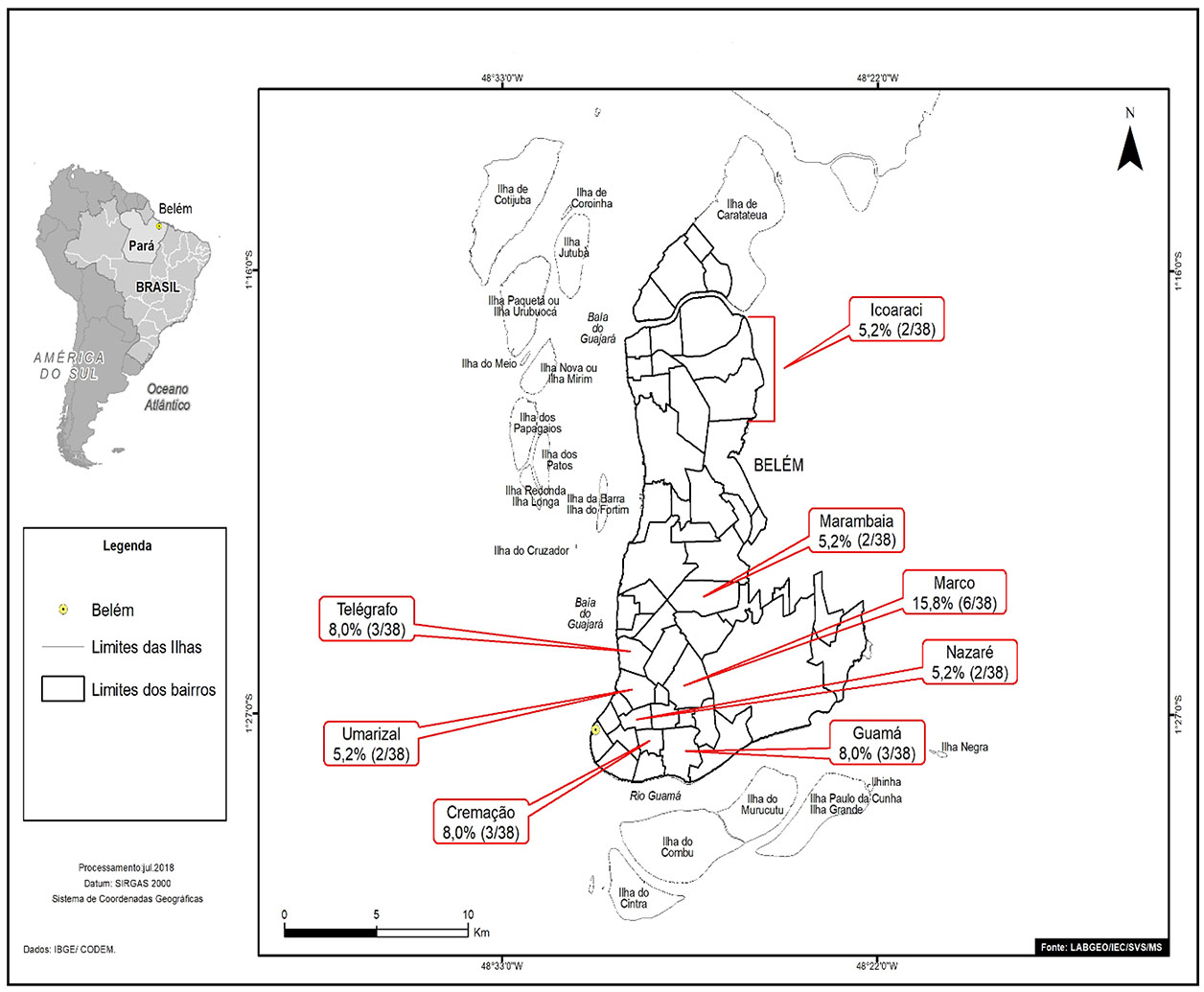
Source: Geoprocessing Laboratory (LabGeo/IEC/SVS/MS).
Figure 1 - Distribution by neighborhoods of Belém, of positive samples for anti-HCV antibodies from patients diagnosed with HNANB treated at SAHEP/IEC, from 1982 to 1988, in Belém, Pará State, Brazil
In the municipality of Ananindeua, most positive samples (3/5; 60.0%) were from patients who lived in Cidade Nova neighborhood (Figure 2).
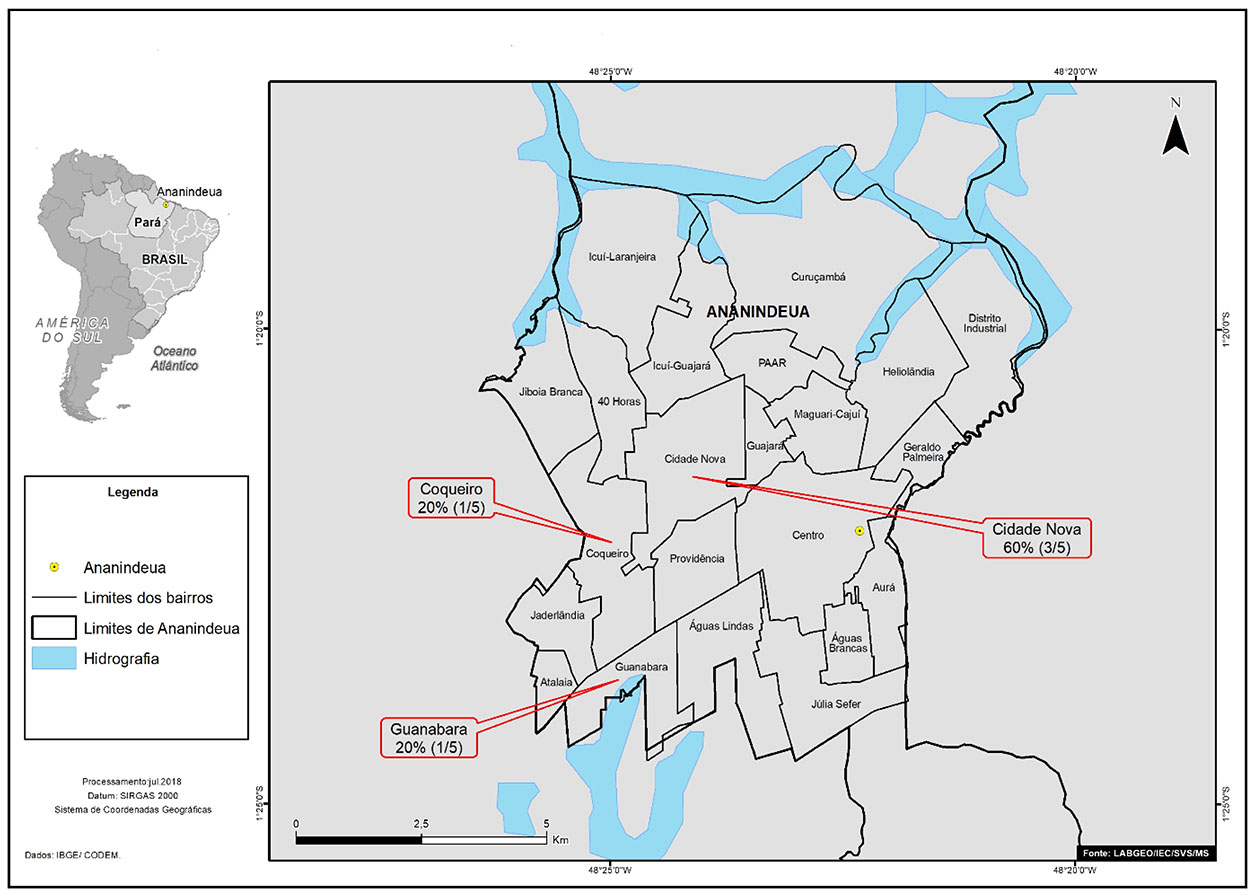
Source: Geoprocessing Laboratory (LabGeo/IEC/SVS/MS).
Figure 2 - Distribution by Ananindeua neighborhoods of positive samples for anti-HCV antibodies of patients diagnosed with HNANB treated at SAHEP/IEC, from 1982 to 1988, in Ananindeua, Pará State, Brazil
Anti-HCV antibodies were detected in 10.9% (43/396) of the samples, with a similar prevalence between genders. The highest frequencies of anti-HCV were observed in adults over 20 and under 60 years of age, especially the age group from 51 to 60 years. There was no statistical significance between sex and age groups when correlated with the presence of HCV antibody. It was possible to detect the presence of viral RNA in 55.8% (24/43) of the samples seroreactive to HCV. For the positive results, according to sex and age groups, there was no statistical significance when correlated with HCV infection (Table 2).
Table 2 - Prevalence of anti-HCV+ and HCV-RNA+, according to sex and age groups, in patients diagnosed with HNANB treated at SAHEP/IEC, from 1982 to 1988, in Belém and Ananindeua, Pará State, Brazil
| Variables | Examined N = 396 | Anti-HCV+ N = 43 | % | p value α = 0,05 | HCV-RNA+ N = 24 | % | p value α = 0,05 |
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Male | 219 | 22 | 10,0 | 0.6775* | 14 | 58,3 | 0,7574† |
| Female | 177 | 21 | 11,9 | 10 | 41,7 | ||
| Age group (years) | |||||||
| ≤ 10 | 63 | 3 | 4,8 | 0,2694‡ | - | - | |
| 11-20 | 75 | 6 | 8,0 | 1 | 4,2 | ||
| 21-30 | 111 | 15 | 13,5 | 9 | 37,5 | ||
| 31-40 | 57 | 8 | 14,0 | 5 | 20,8 | 0,5719‡ | |
| 41-50 | 40 | 3 | 7,5 | 3 | 12,5 | ||
| 51-60 | 23 | 6 | 26,1 | 5 | 20,8 | ||
| ≥ 61 | 27 | 2 | 7,4 | 1 | 4,2 |
N: Sample number; %: Prevalence; Conventional sign used: - Numerical data equal to zero, not resulting from rounding; * χ2 (Yates) =0.173; † χ2 (Yates) = 0.095; ‡ G test for independent samples.
Through genotyping, two genotypes (1 and 3), subtypes 1b and 3a, respectively, were found in eight (33.3%) of 24 RNA detected (Chart 1). The cladogram developed for HCV samples was based on the sequence alignment of the NS5B region (382 bp). Three samples grouped with sequences of genotype 1, subtype 1b, and one grouped with genotype 3, subtype 3a (Figure 3).
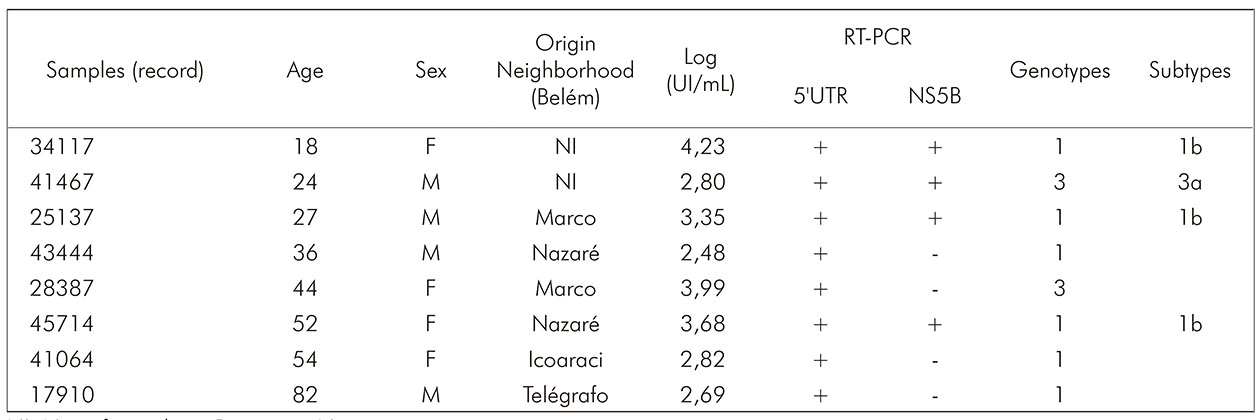
NI: Not informed; +: Positive; -: Negative.
Chart 1 - Demographic characteristics, viral load and HCV genotypes of the eight patients diagnosed with HNANB with detected genotypes, treated at SAHEP/IEC, from 1982 to 1988, in Belém, Pará State, Brazil
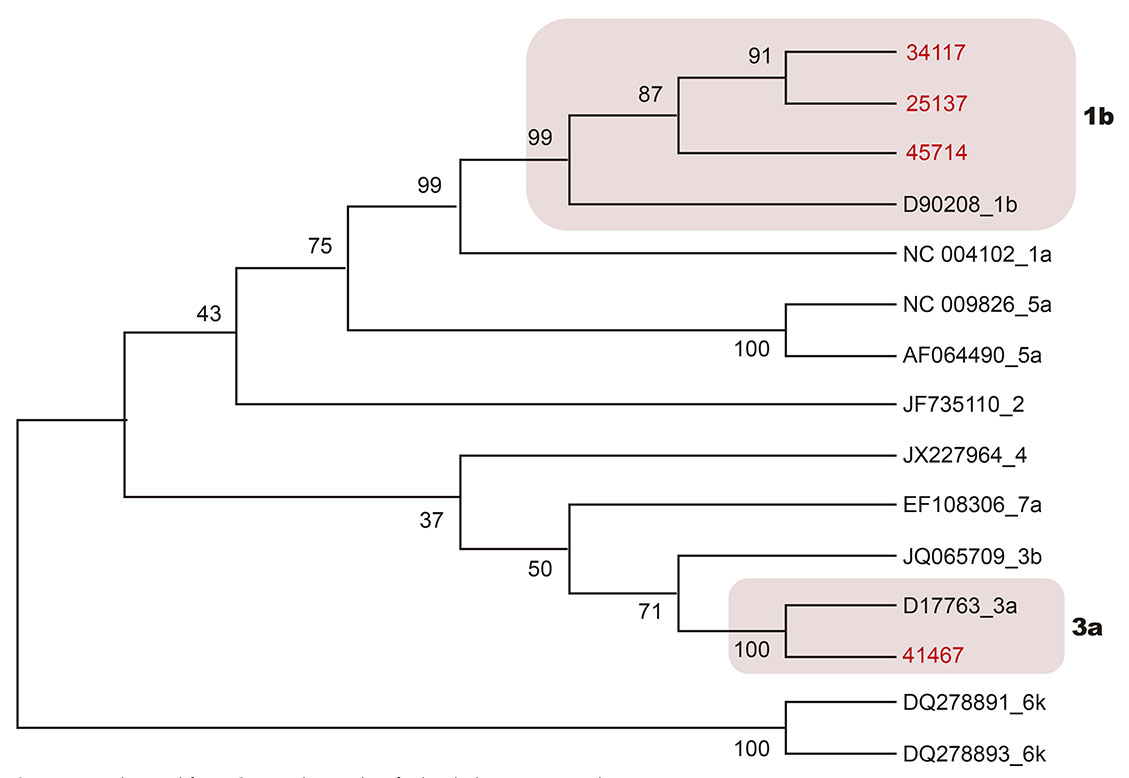
Sequences obtained from GenBank are identified with the access number.
Figure 3 - Cladogram of the NS5B region of HCV inferred with Neighbor-Joining method (Kimura 2-parameters) in the MEGA v7 program
Phylogenetic analysis of the 5'UTR region did not allow the determination of HCV subtypes, because the amplicons of this region presented sequences with a limited number of nucleotide bases (< 230 bp), which made it impossible to infer phylogenetic relationships.
DISCUSSION
Retrospective studies on the prevalence and characterization of HCV are important in order to understand this virus behavior in the population and its dispersion at a time when the specific diagnosis was not available9. Despite the sensitive and advanced molecular methods currently available for the diagnosis of viral hepatitis, the long storage period of the samples, although cryopreserved (-20 ºC), certainly induced changes at the molecular level capable of impairing laboratory analyses, because it is an RNA virus extremely susceptible to degradation, increasing these circumstantial losses15. Nevertheless, this study revealed unprecedented results on the occurrence of hepatitis C in the Brazilian Amazon during the analyzed period (1982 to 1988).
Prior to the discovery of hepatitis C and its etiological agent, the incidence of HNANB in the United States of America in the 1980s, was considered stable, presenting an average of 7.1 cases per 100,000 inhabitants, according to the Centers for Disease Control and Prevention, which for a period of seven years (1982-1988) developed a study in four sentinel municipalities, using samples of patients with HNANB, collected between 1985 and 1988, who were tested for anti-HCV, demonstrating 45% positivity16.
A prospective study conducted in Spain between 1978 and 1984, with 54 patients diagnosed with post-transfusion HNANB, detected 85% of HCV antibodies17. In Paris, between 1980 and 1989, 39.6% of patients submitted to hemodialisise were infected with HCV and this hepatovirus was responsible for 91% of HNANB cases18.
In Brazil, Focaccia et al.19 reported a prevalence of HCV infection of 1.4% in the population of the city of São Paulo. A retrospective study of laboratory reanalysis of HCV infection in samples from 1990 to 1993 in Belém described a prevalence of 62.6% for HCV antibody20. Valois et al.21 observed a 1.1% prevalence for anti-HCV among candidates for blood donation in the state capital.
The seroprevalence for anti-HCV in the sample studied was 10.9%, higher than that observed by Fonseca et al.22, who found positivity of 0.2% of anti-HCV, in a retrospective study conducted in the city of Manaus, State of Amazonas, in the period from 1989 to 1998, in patients diagnosed with acute hepatitis. However, similar to that reported by Teixeira23, who observed positivity of 14.3% of anti-HCV in a research of clinical-laboratory reanalysis of cases reported as HNANB, in the period from 1994 to 1996, also in Belém and Ananindeua.
In the current study, the positivity for anti-HCV was similar between genders, being higher in adults aged between 51 and 60 years. Martins et al.24 suggested that the highest prevalence of hepatitis C, observed after 50 years of age, can be explained by the late diagnosis of HCV exposure.
Regarding sex and age groups, a study conducted by Santos et al.25, with patients diagnosed with HNANB, showed that all individuals reactive to anti-HCV were male, with ages ranging from 27 to 36 years. Simon et al.26, in a research conducted in the city of Paris, France, among hemodialysis patients with HNANB, observed that the mean age of patients with reactive anti-HCV was 50 years, most of them male. Valois et al.21, when analyzing a population of candidates for blood donation, in Belém, they found the male sex as predominant among cases of reactive anti-HCV, with a higher prevalence in the age group between 30 and 39 years.
The professional occupation is a relevant variable in epidemiological studies. In this study, professional occupation was heterogeneous, with the group of students being singled out. Morais and Oliveira27, in a study conducted in southwestern Bahia between 2003 and 2014, reported a prevalence of 0.96% of anti-HCV among students.
In the present study, all samples were from patients of the urban area, from the municipalities of Belém (most of them) and Ananindeua. Marco neighborhood had the highest prevalence of anti-HCV reagent among the neighborhoods of Belém; and Cidade Nova, among the neighborhoods n Ananindeua, which could be explained for being very populous neighborhoods.
Despite the limitations related to the time of storage and conditioning of serum samples, it was possible to detect viral RNA in 55.8% of the 43 seroreagent patients in the present study. Lower results were reported by Fonseca et al.28, who observed a prevalence of 5% of viral RNA detected in the state of Amazonas, with cryopreserved samples collected during the years of 1981 to 1986, from patients with acute HNANB.
In the study by Simon et al.26 conducted in Paris, hemodialysis patients with HNANB, 82.6% (19/23) were positive for HCV RNA.
In Brazil, a retrospective study of laboratory reanalysis of HCV infection in dialysed samples from 1990 to 1993, in the municipality of Belém, described 4.3% of viral RNA detected20.
Oliveira et al.29 observed 62.5% of viral RNA in a study among riverside dwellers in the municipality of Cametá, Pará. Freitas et al.30 reported 5.3% detection for HCV-RNA in patients with anti-HCV in Belém. In the study on the "Epidemiological aspects of HCV infection in users of non-injectable drugs in the state of Pará, eastern Amazonia", there was a prevalence of viral RNA of 28%31.
The HCV-RNA detection data from the present study are similar to that found by Oliveira et al.29, with important emphasis on the fact that the serum samples tested in the mentioned research, unlike those tested in the present series, were newly collected. Variations found in different studies can be explained by the differences between the studied groups and/or cryopreserved samples, and by the time elapsed between the collection and performance of the tests.
In addition to the importance for phylogeny, HCV genotyping is an important factor for the management of infected individuals and for epidemiological purposes that indicates the route of acquisition and affects the clinical outcome and response to treatment32,33. Genotype 1, subtype 1b, is more resistant to certain therapies, and genotype 3, subtype 3a, is significantly associated with the more aggressive forms of the disease32.
HCV genotyping of this study was carried out by sequencing the 5'UTR and NS5B regions. Genotyping was possible in eight of the 24 RNAs detected, most of them were genotyped in the 5'UTR region. Two genotypes (1 and 3) were identified in both regions, in the same serum samples, and two subtypes (1b and 3a) were characterized in the NS5B region.
The 5'UTR is highly conserved; consequently, the annealing of primers is much more conserved in relation to NS5B34. Due to the high level of conservation and sensitivity of 5'UTR, this region has been more used by clinical laboratories for HCV genotyping35. Thus, these characteristics may explain the detection of more genotypes in the 5'UTR region than in the NS5B studied.
It was not possible to discriminate subtypes within the genotypes detected in the 5'UTR region. The 5'UTR region, unlike the NS5B, due to its high level of conservation, does not have sufficient variation to discriminate HCV classifications at the level of viral subtype34,35,36. Thus, the phylogenetic trees of 5'UTR are less-abled to group subtypes of the same genotype into clades than NS5B sequence trees, regardless of the method used for phylogenetic inference34.
HCV genotypes 1 and 3 are the most common cause of infections around the world1,37. In Brazil, genotype 1 is considered the most prevalent cause, followed by genotype 3 and genotype 21,12,37. Among the regions of Brazil, the Northern Region has the highest frequency of genotype 1 (51.7% to 74.1%); the Midwest region, genotype 2 (11.4%); the Southern Region, genotype 3 (43.2%); and in the Southeast Region, in the state of São Paulo, genotypes 4 and 5 were described but rarely found12.
Perone et al.38 conducted a study with patients with chronic hepatitis C, treated at the National Reference Centers for Viral Hepatitis in Belo Horizonte, State of Minas Gerais, between 2002 and 2006, the study observed a high prevalence of genotype 1 (78.4%) and genotype 3 (17.9%). Oliveira et al.39 reported the detection of 77.1% for genotype 1 and 19.82% for genotype 3 in a study conducted in a reference hospital for infectious diseases in the state of Goiás.
It was possible to genotyping one third of the samples positive for HCV-RNA, with predominance of genotype 1, similar to the study by Sawada et al.36, which concluded that, in the distribution of HCV in different exposure categories in Pará, there is a predominance of genotype 1.
The distribution of genotypes identified in the present study was similar to that found by Baia20 who described the detection of 73% for genotype 1 and 24.3% for genotype 3 in a retrospective study among patients on dialysis, from 1990 to 1993, in Belém.
Data from the current study were also similar to findings of other authors in the Northern Region, demonstrating the importance of genotypes 1 and 3 in the epidemiology of this virus: Campiotto et al.12 described the detection of 74.1% for genotype 1 and 24.7% for genotype 3 in the Northern Region; Araújo et al.40 found a prevalence of 76.1% for genotype 1 and 19.6% for genotype 3 in patients with chronic hepatitis in the state of Amazonas; Oliveira et al.39 observed 76.9% of genotype 1 and 23.1% of genotype 3 in non-injectable drug users in Pará; Guimarães et al.41 detected 72.4% of genotype 1 and 23.3% of genotype 3 in the general population, in the state of Pará, and described a prevalence of 76.7% for genotype 1 and 20.7% for genotype 3 in the Metropolitan Region of Belém.
A retrospective study conducted in the Amazon also analyzed cryopreserved serum samples at -70 ºC, collected from 1981 to 1986, for subsequent analysis of HCV28. Another study also retrospective was conducted by Baia20 in samples placed in biobank (-20 ºC) for more than 20 years. These studies corroborate the present research, highlighting the importance of the use of institutional biobanks in health research.
CONCLUSION
Despite the limitations of this research, due to the long storage time of the samples (three decades), it was possible to detect 10.9% of anti-HCV antibodies and characterize HCV in sera from biobank, revealing the circulation of at least two genotypes and two subtypes of the virus: a) genotype 1, detected in most genotyped samples and, currently, exhibiting greater resistance to drugs than other known genotypes; and b) genotype 3, with less frequency in the sample from this study, but currently associated with greater virulence.
It was concluded that the analysis of biobank samples allowed the identification of HCV strains, and may contribute for future studies with the objective of highlighting evolutionary aspects associated with the resistance and virulence of this pathogen.
It is suggested that the institutions evaluate the best way to packaging serum samples, such as freezing at lower temperatures, lyophilization, etc.; as well as the benefit-cost to modernizing procedures in institutional biobanks aimed at health research, due to its relevance for present and future studies.
ACKNOWLEDGMENT
To all those who participated and contributed to this study; to SAHEP/IEC/SVS/MS workers for their collaboration during the development and review of this article.
REFERENCES
1 Polaris Observatory HCV Collaborators. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol. 2017 Mar;2(3):161-76. [ Links ]
2 European Association for the Study of the Liver. EASL recommendations on treatment of hepatitis C 2018. J Hepatol. 2018 Aug;69(2):461-511. [ Links ]
3 Sociedade Brasileira de Hepatologia. Sociedade Brasileira de Infectologia. Consenso. Recomendações das Sociedades Brasileiras de Hepatologia (SBH) e Infectologia (SBI) para o tratamento da hepatite C no Brasil com novos medicamentos antivirais de ação direta (DAAs). Braz J Infect Dis. 2016 mar;20(2 Supl 1):S2-7. [ Links ]
4 Ministério da Saúde (BR). Secretaria de Vigilância em Saúde. Departamento de Vigilância, Prevenção e Controle das IST, do HIV/Aids e das Hepatites Virais. Manual Técnico para o Diagnóstico das Hepatites Virais. 2. ed. Brasília: Ministério da Saúde; 2018. [ Links ]
5 Viana DR, Veloso NM, Carvelho Neto O, Papacota NG, Nunes GM, Guedes VR. Hepatite B e C: diagnóstico e tratamento. Rev Patol Tocantins. 2017 set;4(3):73-9. [ Links ]
6 Pawlotsky JM, Feld JJ, Zeuzem S, Hoofnagle JH. From non-A, non-B hepatitis to hepatitis C virus cure. J Hepatol. 2015 Apr;62(1 Suppl): S87-99. [ Links ]
7 Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989 Apr;244(4902):359-62. [ Links ]
8 International Committee on Taxonomy of Viruses. Virus taxonomy: 2017 release [Internet]. Birmingham (AL): ICTV; 2017 [cited 2017 Jun 17]. Available from: Available from: https://talk.ictvonline.org/taxonomy/ . [ Links ]
9 Conte VP. Hepatite crônica por vírus C. Parte 1: considerações gerais. Arq Gastroenterol. 2000 jul-set;37(3):187-94. [ Links ]
10 Morice Y, Roulot D, Grando V, Stirnemann J, Gault E, Jeantils V, et al. Phylogenetic analyses confirm the high prevalence of hepatitis C virus (HCV) type 4 in the Seine-Saint-Denis district (France) and indicate seven different HCV-4 subtypes linked to two different epidemiological patterns. J Gen Virol. 2001 May;82(Pt 5):1001-12. [ Links ]
11 Sandres-Sauné K, Deny P, Pasquier C, Thibaut V, Duverlie G, Izopet J. Determining hepatitis C genotype by analyzing the sequence of the NS5b region. J Virol Methods. 2003 May;109(2): 187-93. [ Links ]
12 Campiotto S, Pinho JR, Carrilho FJ, Silva LC, Souto FJ, Spinelli V, et al. Geographic distribution of hepatitis C virus genotypes in Brazil. Braz J Med Biol Res. 2005 Jan;38(1):41-9. [ Links ]
13 Ayres M, Ayres Jr M, Ayres DL, Santos AS. BioEstat 5.0: aplicações estatísticas nas áreas das ciências biológicas e médicas. Belém: Sociedade Civil Mamirauá; 2007. 364 p. [ Links ]
14 Brasil. Ministério da Saúde. Resolução nº 466, de 12 de dezembro de 2012. Aprova normas regulamentadoras de pesquisas envolvendo seres humanos. Diário Oficial da União, Brasília (DF), 2013 jun 12; Seção 1:59. [ Links ]
15 Moreau de Gerbehaye AI, Bodéus M, Robert A, Horsmans Y, Goubau P. Stable hepatitis C virus RNA detection by RT-PCR during four days storage. BMC Infect Dis. 2002 Oct;2:22. [ Links ]
16 Alter MJ, Hadler SC, Judson FN, Mares A, Alexander WJ, Hu PY, et al. Risk factors for acute non-A, non-B hepatitis in the United States and association with hepatitis C virus infection. JAMA. 1990 Nov;264(17):2231-5. [ Links ]
17 Esteban JI, Viladomiu L, Gonzalez A, Roget M, Genescà J, Esteban R, et al. Hepatitis C virus antibodies among risk groups in Spain. Lancet. 1989 Aug;334(8658):294-7. [ Links ]
18 Alter MJ. HCV routes of transmission: what goes around comes around. Semin Liver Dis. 2011 Nov;31(4):340-6. [ Links ]
19 Focaccia R, Conceição OJ, Sette H Jr, Sabino E, Bassit L, Nitrini DR, et al. Estimated prevalence of viral hepatitis in the general population of the Municipality of São Paulo, measured by a serologic survey of a stratified, randomized and residence-based population. Braz J Infect Dis. 1998 Dec;2(6):269-84. [ Links ]
20 Baia KSM. Reavaliação laboratorial da infecção pelo vírus da hepatite C em dialisados atendidos no período de 1990-1993, Belém, Pará, Brasil [Monografia]. Belém (PA): Universidade Federal do Pará, Faculdade de Medicina e Cirurgia do Pará; 2013. 58 p. [ Links ]
21 Valois RC, Maradei-Pereira LMC, Crescente JAB, Oliveira-Filho AB, Lemos JAR. VHC infection through perforating and cutting material among candidates for blood donation in Belém, Brazilian Amazon. Rev Inst Med Trop S Paulo. 2014 Nov-Dec;56(6):511-5. [ Links ]
22 Fonseca JCF, Brasil LM. Infecção pelo vírus da hepatite C na região Amazônica brasileira. Rev Soc Bras Med Trop. 2004;37(Supl 2):1-8. [ Links ]
23 Teixeira LSC. Hepatite não-A, não-B: reavaliação clínico-laboratorial de casos notificados nos municípios de Belém e Ananindeua, estado do Pará [Monografia]. Belém (PA): Universidade Federal do Pará, Centro de Ciências da Saúde; 1997. 81 p. [ Links ]
24 Martins T, Narciso-Schiavon JL, Schiavon LL. Epidemiologia da infecção pelo vírus da hepatite C. Rev Assoc Med Bras. 2011 jan-fev;57(1):107-12. [ Links ]
25 Santos A, Carvalho A, Bento D, Sá R, Tomaz J, Rodrigues V, et al. Epidemiologia da hepatite C na região centro de Portugal - prevalência do anti-VHC na população do Distrito de Coimbra. Acta Med Port. 1993;6:567-72. [ Links ]
26 Simon N, Couroucé AM, Lemarrec N, Trépo C, Ducamp S. A twelve year natural history of hepatitis C virus infection in hemodialyzed patients. Kidney Int. 1994 Aug;46(2):504-11. [ Links ]
27 Morais MTM, Oliveira TJ. Perfil epidemiológico e sóciodemográfico de portadores de hepatite C de um município do sudoeste baiano. Rev Saude.Com. 2015 jun;11(2):137-46. [ Links ]
28 Fonseca JCF, Brasil L, Fay F, Castilho M, Botelho R, Borborema C, et al. Prevalência da infecção pelo vírus da hepatite C (VHC) e da hepatite E (VHE) em pacientes com hepatite aguda N-A, N-B, N-D. Arch Argent Enferm Apar Dig. 1996;10:90. [ Links ]
29 Oliveira CSF, Silva AV, Santos KN, Fecury AA, Almeida MKC, Fernandes AP, et al. Infecção pelo vírus da hepatite B e C em ribeirinhos da Amazônia brasileira. Rev Soc Bras Med Trop. 2011 set-out;44(5):546-50. [ Links ]
30 Freitas MJR, Fecury AA, Almeida MKC, Freitas AS, Guimarães VS, Silva AM, et al. Prevalence of hepatitis C virus infection and genotypes in patient with chronic kidney disease undergoing hemodialysis. J Med Virol. 2013 Oct;85(10):1741-5. [ Links ]
31 Oliveira-Filho AB, Sawada L, Pinto LC, Locks D, Bahia SL, Castro JA, et al. Epidemiological aspects of HCV infection in non-injecting drug users in the Brazilian state of Pará, eastern Amazon. Virol J. 2014 Feb;11(38). [ Links ]
32 Kermani FR, Sharifi Z, Ferdowsian F, Paz Z, Zamanian M. Distribution of hepatitis C virus genotypes among chronic infected injecting drug users in Tehran, Iran. Jundishapur J Microbiol. 2013 Feb;6(3):265-8. [ Links ]
33 Moosavy SH, Davoodian P, Nazarnezhad MA, Nejatizaheh A, Eftekhar E, Mahboobi H. Epidemiology, transmission, diagnosis, and outcome of hepatitis C virus infection. Electron Physician. 2017 Oct;9(10):5646-56. [ Links ]
34 Hraber PT, Fischer W, Bruno WJ, Leitner T, Kuiken C. Comparative analysis of hepatitis C virus phylogenies from coding and non-coding regions: the 5' untranslated region (UTR) fails to classify subtypes. Virol J. 2006 Dec;3:103. [ Links ]
35 Murphy DG, Willems B, Deschênes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5' untranslated region sequences. J Clin Microbiol. 2007 Apr;45(4):1102-12. [ Links ]
36 Sawada L, Pinheiro AC, Locks D, Pimenta AS, Rezende PR, Crespo DM, et al. Distribution of hepatitis C virus genotypes among different exposure categories in the State of Pará, Brazilian Amazon. Rev Soc Bras Med Trop. 2011 Jan-Feb;44(1):8-12. [ Links ]
37 Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: an up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016 Sep;22(34):7824-40. [ Links ]
38 Perone C, Del Castillo DM, Pereira GL, Carvalho NO, Januário JN, Teixeira R. Alta prevalência do genótipo 1 em portadores de hepatite C crônica em Belo Horizonte, MG. Rev Soc Bras Med Trop. 2008 mai-jun;41(3):238-42. [ Links ]
39 Oliveira TJB, Reis LAP, Barreto LSLO, Gomes JG, Manrique EJC. Perfil epidemiológico dos casos de hepatite C em um hospital de referência em doenças infectocontagiosas no estado de Goiás, Brasil. Rev Pan-Amaz Saude. 2018 mar;9(1): 51-7. [ Links ]
40 Araújo AR, Almeida CM, Fraporti L, Garcia N, Lima TA, Maia LP, et al. Caracterização do vírus da hepatite C em pacientes com hepatite crônica: genótipos no Estado do Amazonas, Brasil. Rev Soc Bras Med Trop. 2011 set-out;44(5):638-40. [ Links ]
41 Guimarães VS, Melo TG, Ferreira RCD, Almeida SF, Martins LC. Prevalence of hepatitis C virus genotypes in the State of Pará, Brazil. Rev Soc Bras Med Trop. 2018 Jul-Aug;51(4): 508-12. [ Links ]
How to cite this article / Como citar este artigo: Barbosa KMV, Moreira LVL, Oliveira CMA, Souza AJS, Nunes HM, Soares MCP, et al. Hepatitis C in the 1980s: case review of former non-A and non-B hepatitis from a hepatology service in the Brazilian Amazon. Rev Pan Amaz Saude. 2019;10:e201900096. Doi: http://dx.doi.org/10.5123/S2176-6223201900096.
Received: September 28, 2018; Accepted: April 08, 2019










 text in
text in