INTRODUCTION
Human alphaherpesvirus 1 and 2 (HHV-1 and HHV-2), also known as herpes simplex viruses 1 and 2 (HSV-1 and HSV-2), belong to the Herpesviridae family, Alphaherpesvirinae subfamily, genus Simplexvirus. These viruses are ubiquitous, and it is estimated that about 67% and 13% of the population are infected with HHV-1 and HHV-2, respectively1,2. While primary infections are generally self-limited, a disease caused by reactivation can be particularly severe in immunocompromised individuals2. The increased prevalence and survival of immunosuppressed individuals, regardless of whether their immune suppression is due to bone marrow or solid organ transplantation, HIV infection, or antineoplastic treatments, has led to a rising incidence of HHV reactivation and consequent morbidity and mortality2,3.
Acyclovir (ACV) [9- (2-hydroxyethyl methyl) guanine] is a deoxyguanosine analog that is activated when converted to its monophosphate form by viral thymidine kinase (TK) and then to ACV triphosphate by cellular enzymes. It is the gold-standard treatment for HHV-1 and 2 infections. However, HHV strains resistant to ACV and other antiviral drugs are in circulation4,5. ACV resistance results from mutations of the viral TK gene and less often from mutations of the viral DNA polymerase gene4,5. The growing prevalence of ACV-resistant strains is mainly due to the increased number of patients using ACV prophylactically or undergoing treatment of active infections4. The selection of resistant strains is promoted further by the increased use of the ACV-derived TK-dependent drugs valacyclovir and penciclovir5,6.
Information on the epidemiology and drug resistance of HHV isolates in the general population of Brazil is scarce. A cross-sectional study including subjects from the general population carried out in the cities of Fortaleza, Rio de Janeiro, Porto Alegre, and Manaus showed that the seroprevalences of HHV-1 and HHV-2 antibodies were 67.2% and 11.3%, respectively, without sex differences, and were higher in the North Region7. A seroepidemiological study carried out in Rio de Janeiro from March 1996 to March 1997 among male individuals who had sex with other men showed that the prevalence of HSV-1 and HSV-2 was 83.5% and 63.4%, respectively8. A recent study showed that the HSV-1 and HSV-2 prevalence among pregnant women in Rio de Janeiro was 80.2% and 12.5%, respectively9. In Londrina, Paraná State, a study analyzed five HHV isolates collected between June 2009 and June 2010 from clinical specimens of individuals with either oral or genital lesions, clinically compatible with herpesvirus infection, and demonstrated that all the isolates were sensitive to ACV10. Therefore, the present study was designed to isolate and characterize HHV strains circulating among healthy individuals and to determine their ACV susceptibility.
MATERIALS AND METHODS
Samples were collected by convenience sampling. To recruit volunteers, posters were distributed at the Universidade Federal do Rio de Janeiro campuses in Rio de Janeiro, Brazil. All volunteers who agreed to participate and stated that they did not have an immunocompromise history or chronic diseases were included in the study. No professional health survey was performed. Between October 2008 to September 2010, 15 samples from lesions suggestive of herpetic infections were obtained from 15 healthy individuals who volunteered to participate in the study. All patients reported previous herpetic lesions. However, detailed information on previous diagnostic confirmation, number or frequency of episodes was not provided. The specimens were collected from mucosal or cutaneous lesions (oral, genital, thigh, or torso) of healthy HHV-infected subjects using sterile swabs that were drained into tubes containing 2.5 mL of the viral transport medium (VTM) Gey's Balanced Salt Solution plus 200 U/mL penicillin and streptomycin (Life Technologies, Carlsbad, CA, USA), 2.5 mg/mL amphotericin B (Life Technologies). The samples in VTM were stored in nitrogen until process. None of the study subjects were undergoing ACV prophylaxis or treatment at the time of sample collection. Vero cells were obtained from the Banco de Células do Estado do Rio de Janeiro and maintained in Dulbecco's modified Eagle medium (DMEM, Life Technologies) supplemented with 200 U/mL penicillin and streptomycin, 2.5 mg/mL amphotericin B, and 10% heat-inactivated fetal bovine serum (FBS, Life Technologies). Cell cultures were maintained at 37 ºC in a 5% CO2 environment.
To propagate the viral isolates, cell monolayers were prepared in 24-well plates and the samples were inoculated in duplicate and incubated for 1 h at 37 ºC. A 0.1 mL aliquot of clinical material in VTM was inoculated per well. Wells were subsequently washed with fresh DMEM without FBS. Infected cells were cultured as described above and monitored daily for the event of viral cytopathic effect (CPE) using an inverted microscope. When approximately 75% of the cells showed signs of CPE, or after seven days of incubation without CPE, the cell cultures were submitted to a freezing/thaw cycle and then stored at -70 ºC until further use.
Viral isolates were identified by amplification and sequencing of a 601 bp fragment of the viral DNA polymerase gene using specific primers for HHV-1/211. Briefly, nucleic acids were extracted from 200 µL mixtures of supernatant and cell lysates (viral suspension) employing a Wizard® Genomic DNA Purification KIT (Promega, Madison, WI, USA) per the manufacture's recommendations. A polymerase chain reaction (PCR) was meted out using primers HHV-F1 and HHV-R1 as previously described11. PCR reactions consisted of 5 µL extracted DNA mixed with 45 µL PCR mix containing 0.5 µM each primer and 1x PCR buffer, 2 mM MgCl2, 0.4 µM deoxyribonucleotide triphosphates, and 1 U Taq DNA polymerase (Ludwig Biotec, Alvorada, RS, Brazil). PCR mixtures were subjected to the subsequent amplification cycle conditions: 94 ºC for 3 min followed by 35 cycles at 94 ºC for 45 s, 65 ºC for 1 min, 72 ºC for 1 min, and a final extension at 72 ºC for 7 min. PCR products were visualized by subjecting PCR reactions to 1.2% agarose gel electrophoresis followed by ethidium bromide staining.
Amplification products were sent to Hellixa Genomics Service Provider (Paulínea, São Paulo, Brazil) for sequencing. Overlapping sequences were assembled and edited using SeqMan, EditSeq, and MegAlign within the Lasergene software package (DNASTAR). Phylogenetic analysis was performed with MEGA software v.7.0.1412. A dendrogram was constructed using the Maximum Likelihood method supported the Kimura 2-parameter model. Statistical significance was estimated by bootstrap analysis with 1,000 pseudoreplicates. Sequences were compared to those of HHV-1 and HHV-2 strains obtained from GenBank® (https://www.ncbi.nlm.nih.gov/nucleotide/). Sequences generated for HHV strains analyzed during this study were deposited into GenBank under accession numbers KX650157-KX650171. Nucleotide identities were determined by using the MegAlign p-distance algorithm.
Viral suspensions were titrated to quantify viral infectivity by the endpoint dilution method. Twenty-five microliters of logarithmic dilutions of viral suspensions were used to inoculate confluent Vero cell monolayers in 96-well plates (six wells/dilution). Titers were calculated according to the endpoint dilution assay and expressed as log10 TCID50 (the amount of virus which will produce morphological change in 50% of inoculated cells/25 µL)13.
ACV susceptibility (Sigma-Adlrich, St. Louis, MO, USA) was assessed by comparing viral titer reductions within the presence of serial dilutions of ACV in Vero cell cultures so as to work out the EC50 (effective concentration that reduces in 50% the viral replication)14. Sensitivity patterns were defined by the subsequent values of EC50: sensitive, < 4 μM; intermediate, ≥ 4 μM - ≤ 8,9 μM; resistant, ≥ 9 μM4,14.
This study was approved by the Research Ethics Committee of the Hospital Universitário Clementino Fraga Filho, Universidade Federal of Rio de Janeiro, Rio de Janeiro, Brazil (process number 019/08).
RESULTS
The 15 samples were collected from lesions in genitalia (n = 3), thigh (n = 2), torso (n = 1), and orolabial region (n = 9) (Table 1, Figure 1). HHV was isolated from all 15 samples. Sequence analysis of the DNA polymerase gene showed that 10 isolates belonged to the species HHV-1 and five to HHV-2 (Table 1 , Figure 2). Nine HHV-1 isolates were obtained from lesions located above the waist and one isolate was obtained from a thigh lesion. Conversely, four HHV-2 isolates were obtained from lesions located below the waist (genitalia and thigh) and one was isolated from an orolabial lesion (Table 1).
Table 1 - Characteristics of viral strains
| Samples | Site of collection | Virus species | ACV susceptibility (EC50 3, µM) |
|---|---|---|---|
| ACL | Orolabial | HHV-1 | Sensitive (< 2) |
| ACR | Orolabial | HHV-1 | Sensitive (< 2) |
| APL | Torso | HHV-1 | Resistant (9.48) |
| DPL | Orolabial | HHV-1 | Intermediate (4.24) |
| GA | Orolabial | HHV-1 | Sensitive (< 2) |
| GAB | Orolabial | HHV-1 | Sensitive (< 2) |
| GSM | Orolabial | HHV-1 | Sensitive (< 2) |
| LAF | Orolabial | HHV-1 | Sensitive (< 2) |
| RIC | Orolabial | HHV-1 | Sensitive (< 2) |
| RV | Thigh | HHV-1 | Sensitive (< 2) |
| ADL | Orolabial | HHV-2 | Sensitive (< 2) |
| AM | Thigh | HHV-2 | Sensitive (3.07) |
| EP | Genitalia | HHV-2 | Sensitive (< 2) |
| JLS | Genitalia | HHV-2 | Sensitive (< 2) |
| RO | Genitalia | HHV-2 | Sensitive (< 2) |
HHV-1: Human herpesviruses 1; HHV-2: Human herpesviruses 2; ACV: Acyclovir; EC50: Effective concentration that reduces in 50% the viral replication.
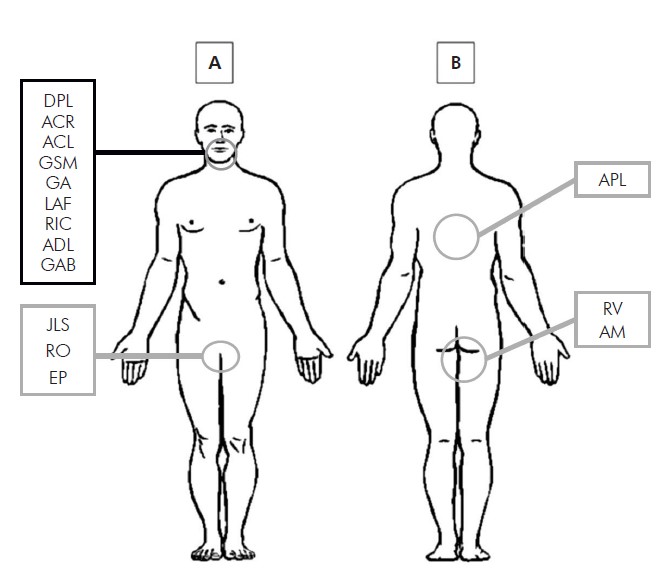
A: Orofacial region and oral cavity: DPL, ACR, ACL, GSM, GA, LAF, RIC, ADL, GAB; Genital region: JLS, RO, EP. B: Trunk and thigh region: APL, RV, AM.
Figure 1 - Illustrative diagram of the sample collection body area
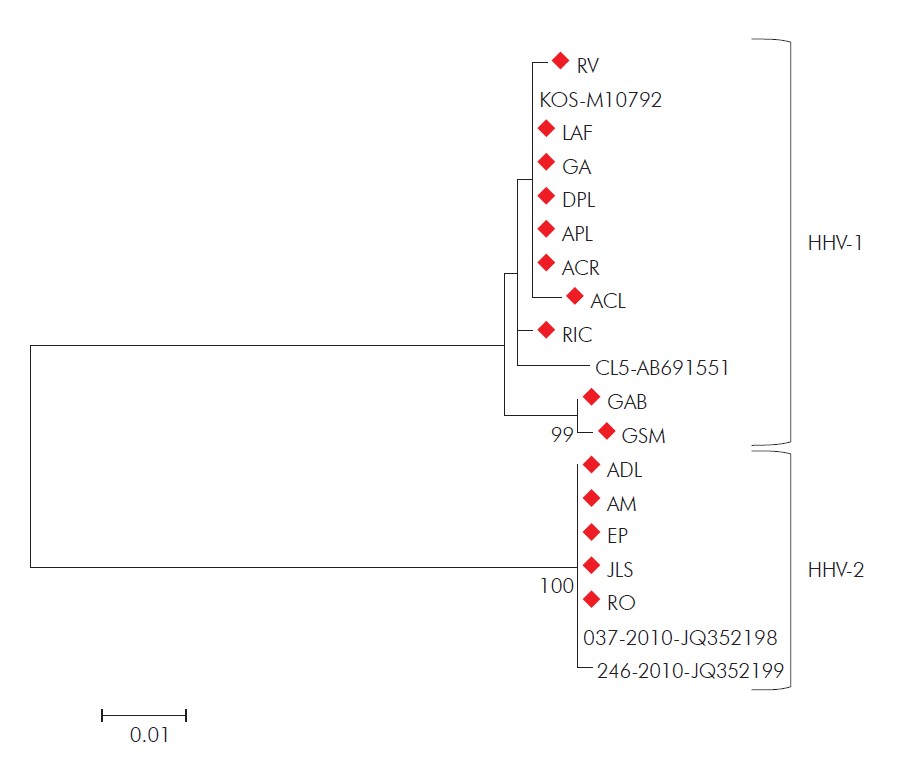
Distances were corrected using the Kimura 2-parameter model, and the dendrogram was constructed using the maximum likelihood method. Statistical support was provided by bootstrapping 1,000 pseudoreplicates. Bootstrap values above 75% are given as branch nodes. Genbank reference strain accession numbers are shown next to the strain identification. Strains detected in this study are indicated by red diamonds.
Figure 2 - Dendrogram constructed from the partial sequences of the gene encoding HHV DNA polymerase of strains isolated in this study
ACV susceptibility was determined for all isolates and for reference strains known to be sensitive or resistant. Results were plotted on a dose-response curve allowing EC50 value calculations (Table 1). Of the 10 HHV-1 strains, one was ACV-resistant, and one showed intermediate sensitivity. All HHV-2 strains were ACV-sensitive.
DISCUSSION
The prevalence of ACV-resistant HHV-1 and HHV-2 strains among otherwise healthy individuals diagnosed with recurrent genital herpes, encephalitis, or disseminated infections was low (< 1%)4,5. However, a higher prevalence (6.4%) of ACV-resistant HHV-1 isolates has been reported among healthy patients with herpetic keratitis15, and some of these cases have been refractory to ACV therapy16,17,18,19. On the other hand, the estimated prevalence in immunocompromised individuals is 3.5% to 14%, reaching 36% depending on the type of immunosuppression4,5,19. The present study analyzed samples from healthy patients with recurrent HHV infections not undergoing antiviral therapy at the time of sample collection and found two HHV strains with either resistance or intermediate sensitivity to ACV. In contrast, the only study on the subject previously published in Brazil did not report the occurrence of ACV-resistant strains10.
Two surveillance studies conducted in France have shown contrasting data regarding the prevalence of antiviral drug resistance among HHV-1 and HHV-2 isolates. A three year surveillance study analyzed 3,900 HHV strains and found no statistical difference in ACV resistance between HHV-1 and HHV-2 strains6. In contrast, a four year surveillance analysis of 144 HHV strains found a significantly higher prevalence of resistance among HHV-2 than HHV-1 isolates16. In this study, ACV resistance was observed only among HHV-1 strains.
Differences in the prevalence of ACV-resistant strains among healthy and immunocompromised patients could be due to two causes: i) viral replication may be prolonged in immunocompromised patients, leading to true persistence; or ii) impaired host immune responses allow less pathogenic viruses incapable of replicating in a healthy host to survive4. However, the factors associated with developing resistance among healthy hosts are incompletely understood. Prolonged ACV prophylaxis and concurrent long-term steroid therapy may promote the emergence of resistance in otherwise healthy patients with recurrent herpetic keratitis17 or genital herpes20, especially if ACV dosing is suboptimal20. Steroid therapy facilitates the establishment of transient immunosuppression, while sub-inhibitory drug concentrations permit viral replication and apply selective pressure. In the present study, patients presented with recurrent infections, and none were undergoing steroid treatment or antiviral therapy at the time of sample collection. Therefore, the development of resistance by this isolate is challenging to explain. One possibility is that a drug-resistant strain caused the primary infection. However, this is just a purely speculative explanation since no data were found in the literature supporting it.
A limitation of this study was the lack of detailed information about the volunteers' history, particularly regarding the number and frequency of recurrence of lesions. This information would be relevant to try to relate lesions' recurrence with the emergence of drug resistance. A previous study evaluated the ACV susceptibility and genetic characterization of sequential corneal HSV-1 isolates in immunocompetent patients. It demonstrated that ACV susceptibility of sequential isolates changed in eight of 15 patients, from ACV-sensitive to ACV-resistant (five patients) and ACV-resistant to ACV-sensitive (three patients)21.
Management of patients infected with ACV-resistant strains requires special attention. However, the same attention level should be given to those infected with intermediate-sensitive strains because the selection of ACV resistance may occur within hours or days after the initiation of treatment6,22. Sensitive and resistant variants co-infections facilitate the rapid selection of resistant strains, especially if treated inadequately22,23.
Taking these observations into account, determining the drug sensitivity of HHV clinical isolates is critical to establish appropriate treatment protocols. Cases caused by resistant or intermediate-resistant strains can be treated with TK-independent antiviral agents such as foscarnet, cidofovir, or brincidofovir. However, these drugs must be administered with caution because ACV-resistant strains may acquire foscarnet resistance under drug selection pressure. Nevertheless, alternating antiviral treatments remain the best strategy for minimizing the emergence of resistant strains6.
A notable finding of this study was the correlation between viral species and the anatomic location of the source lesion. Previously, HHV-1 infections were associated with lesions above the waist, while those caused by HHV-2 occurred below the waist. However, HHV-2 infections above the waist have been increasing. These alterations in anatomic distribution may be due to changes in sexual behavior, with increased oral-genital contact between partners6,24,25. The results demonstrated one HHV-2 strain isolated from an orofacial lesion and one HHV-1 strain from a thigh lesion.
CONCLUSION
The results presented in this study demonstrate the circulation of HHV-1 and HHV-2 in healthy individuals in Rio de Janeiro and highlight the need for continued surveillance of the emergence of HHV strains resistant to antiviral drugs, especially to ACV, a first-line therapeutic agent. The circulation of resistant strains among healthy individuals is of major concern as these individuals represent a potential reservoir of transmission to the general population and immunocompromised patients at greater risk of developing severe and disseminated infections. Identifying ACV resistance is essential
to inform/redirect treatments, monitor therapeutic response, determine optimal drug concentration breakpoints for susceptibility, and further understand the resistance mechanisms.